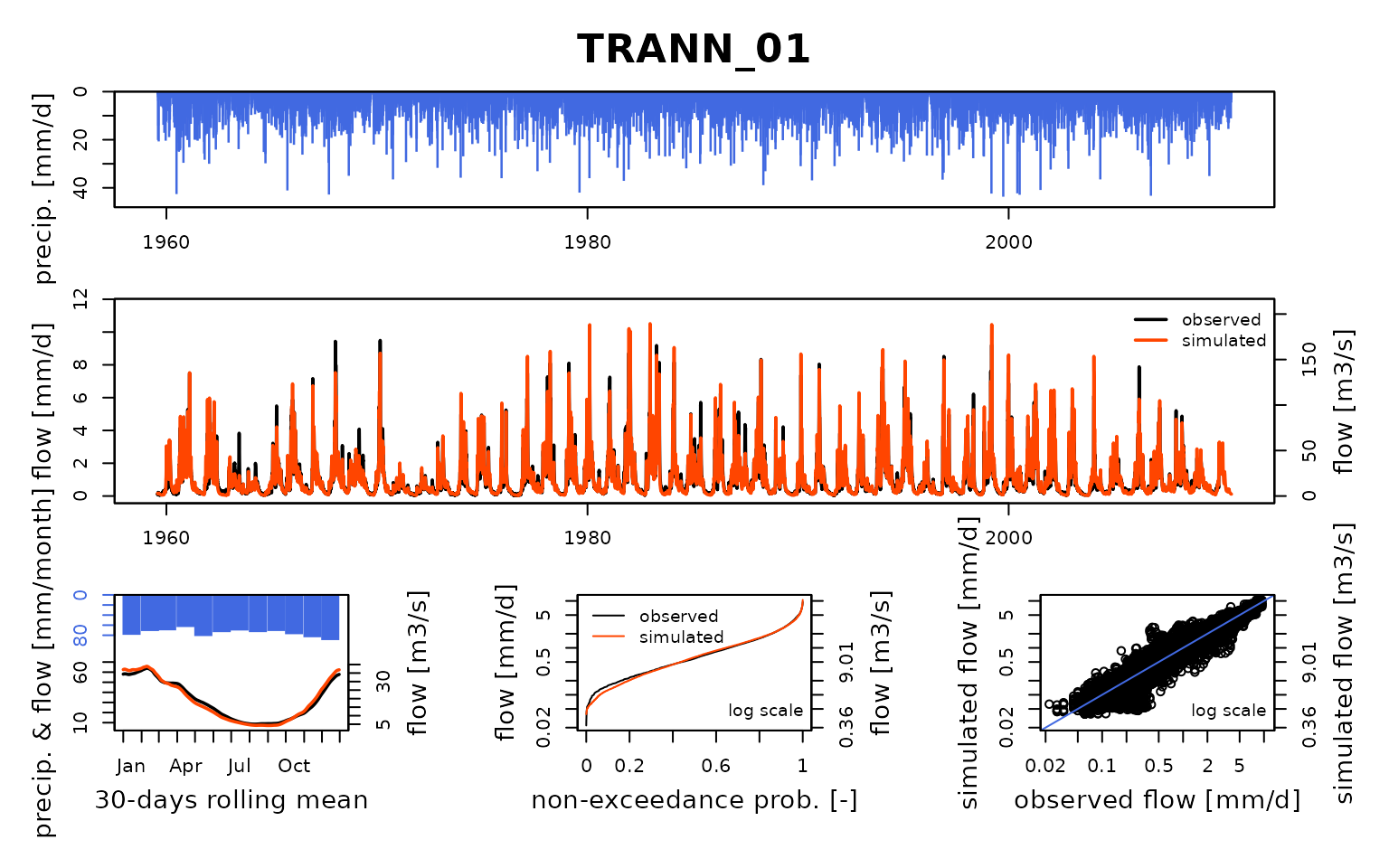
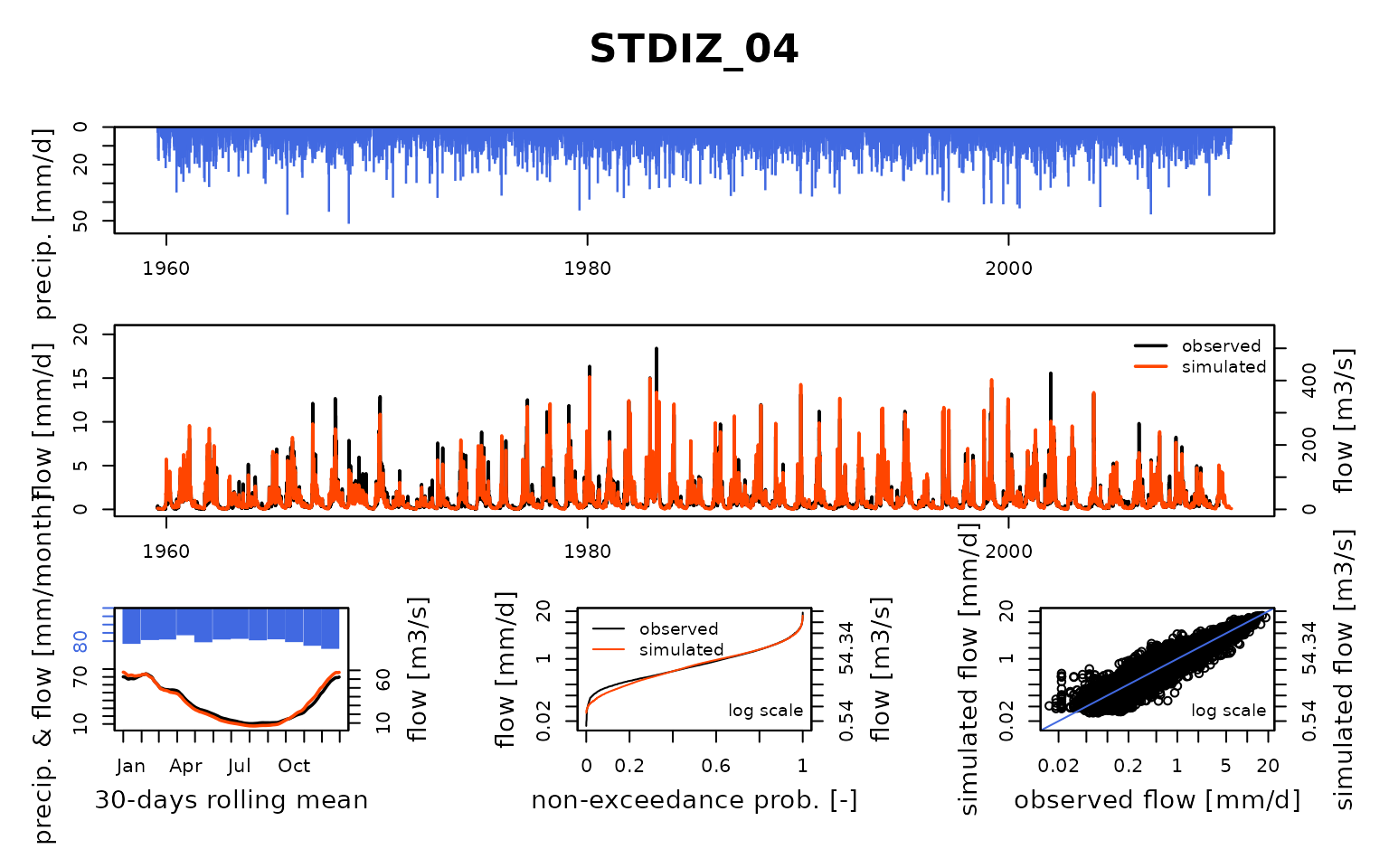
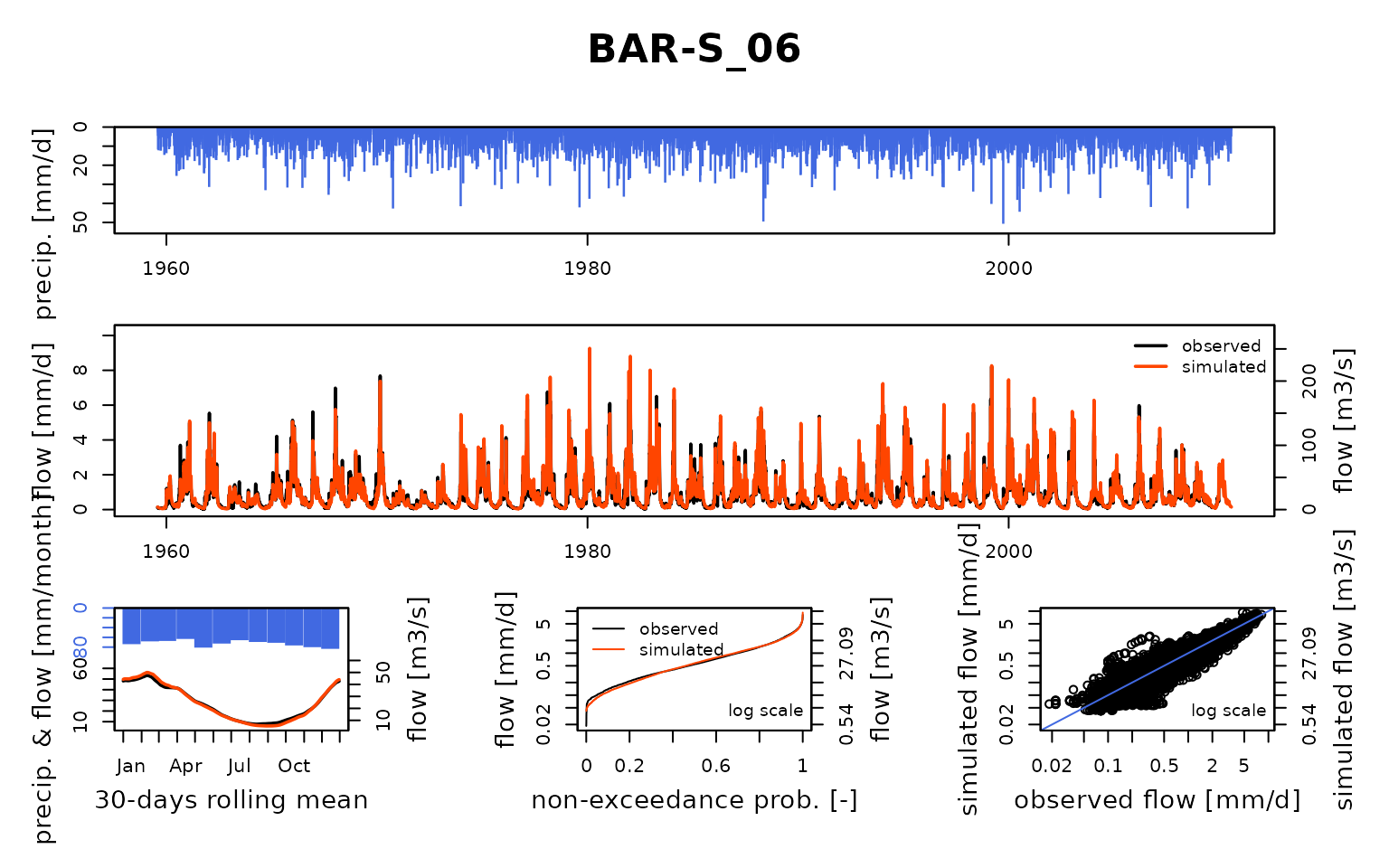
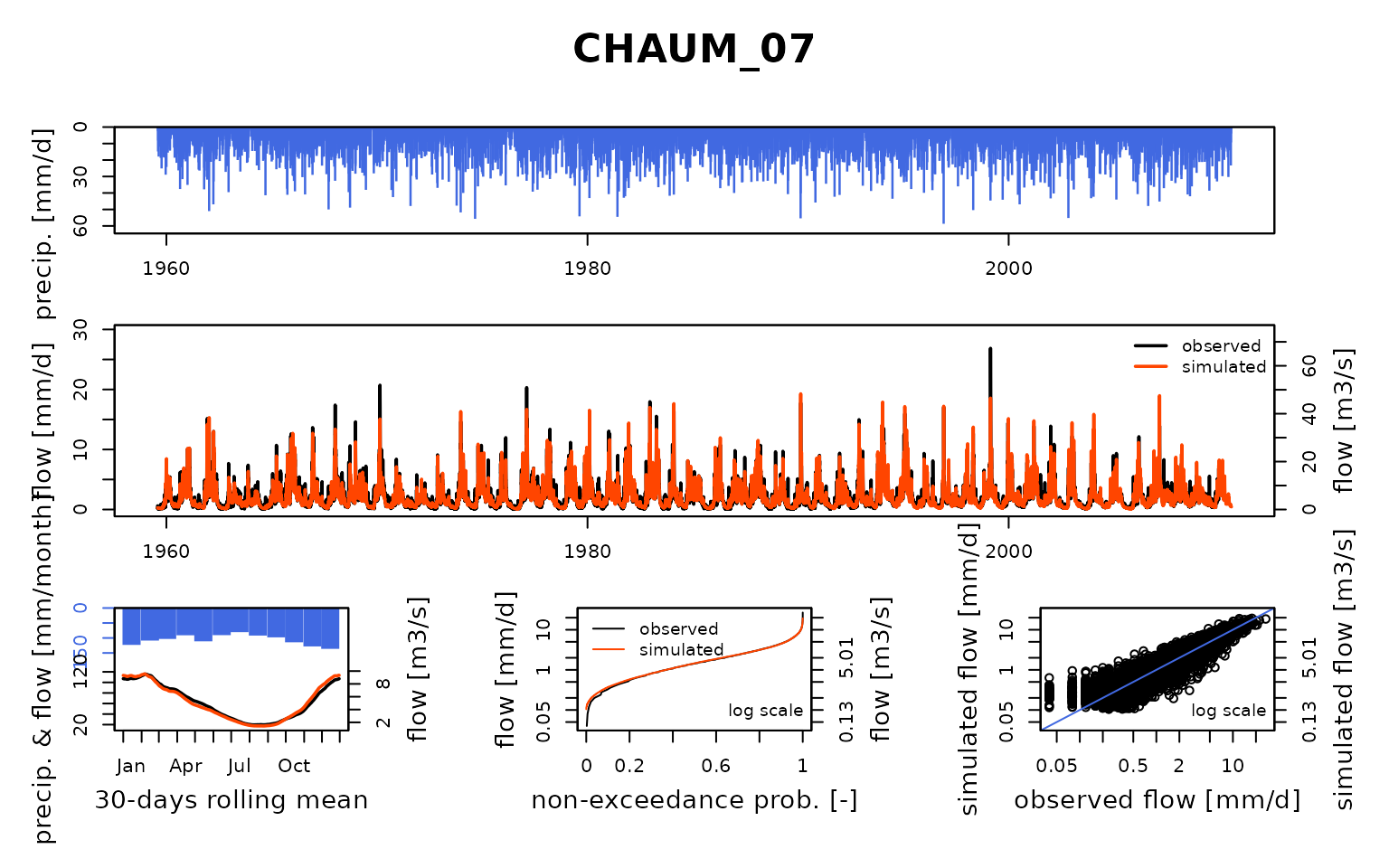
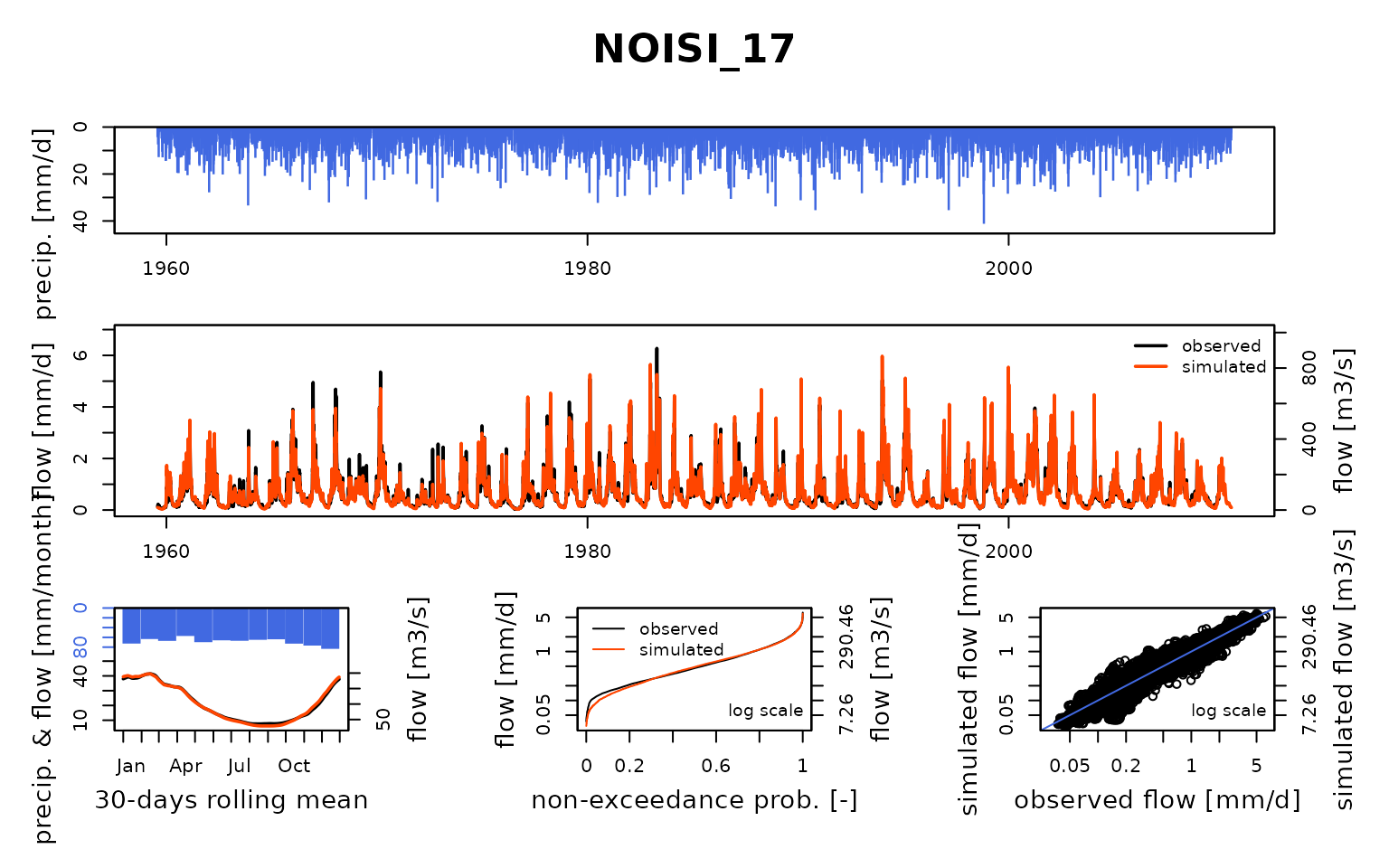
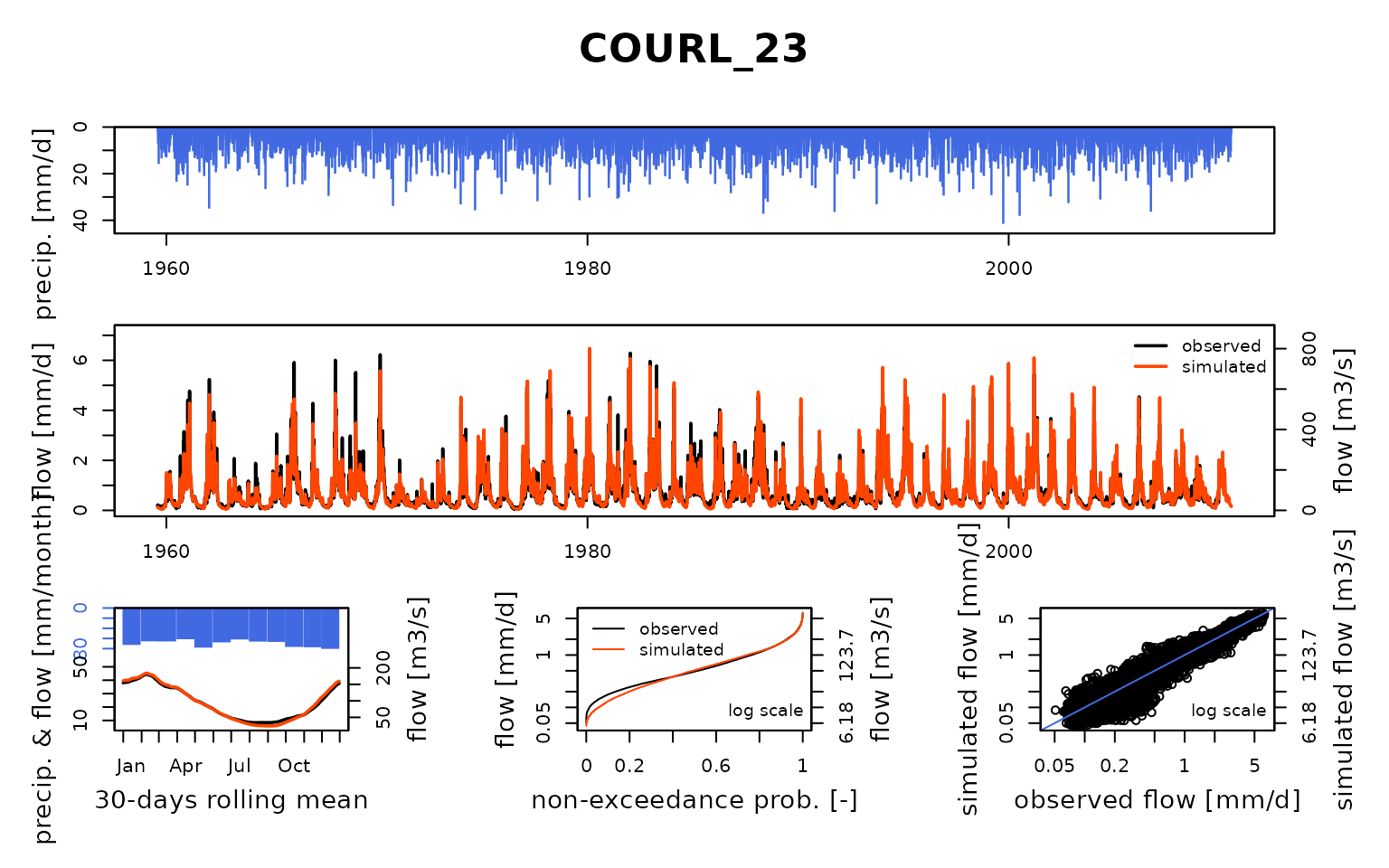
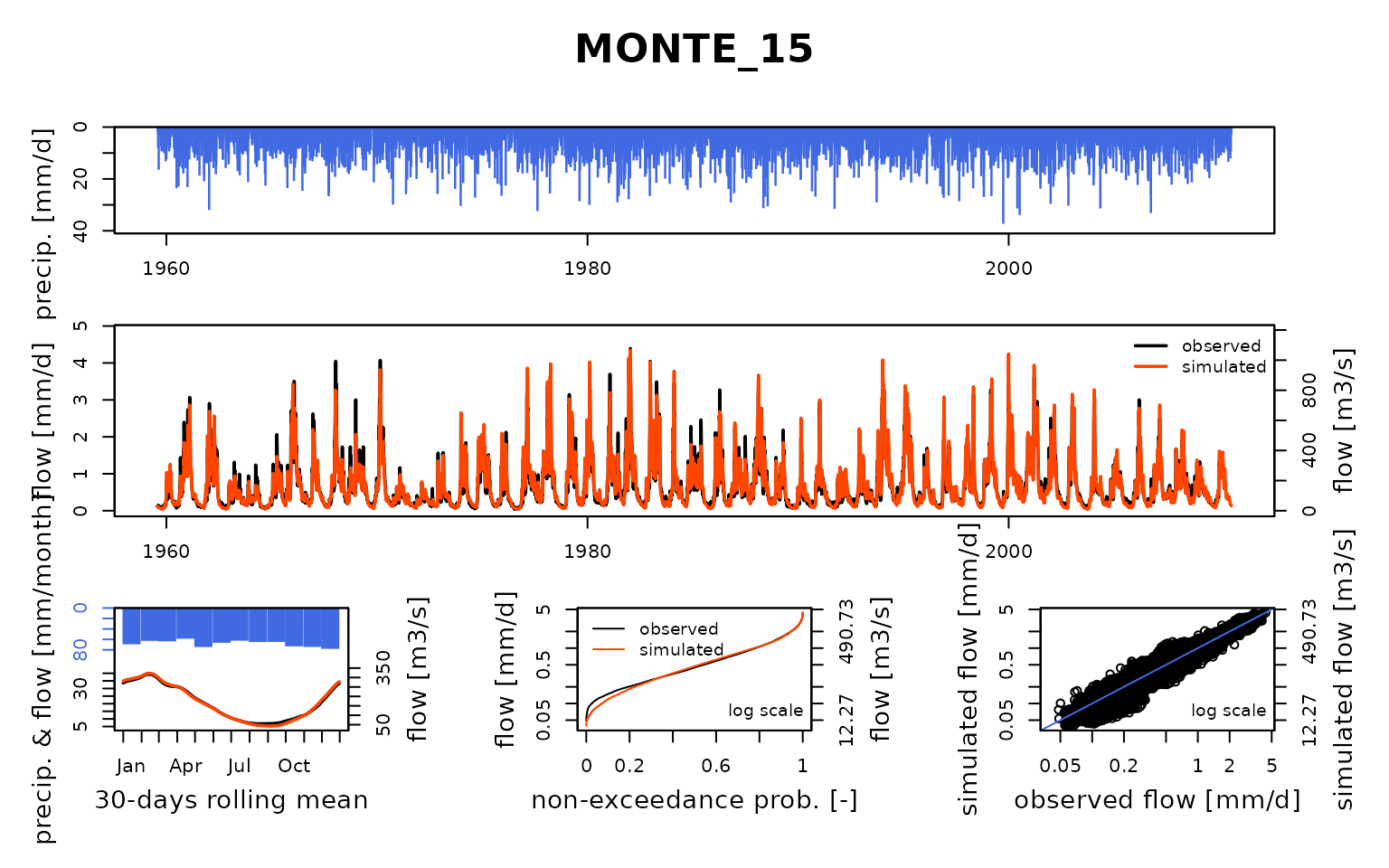
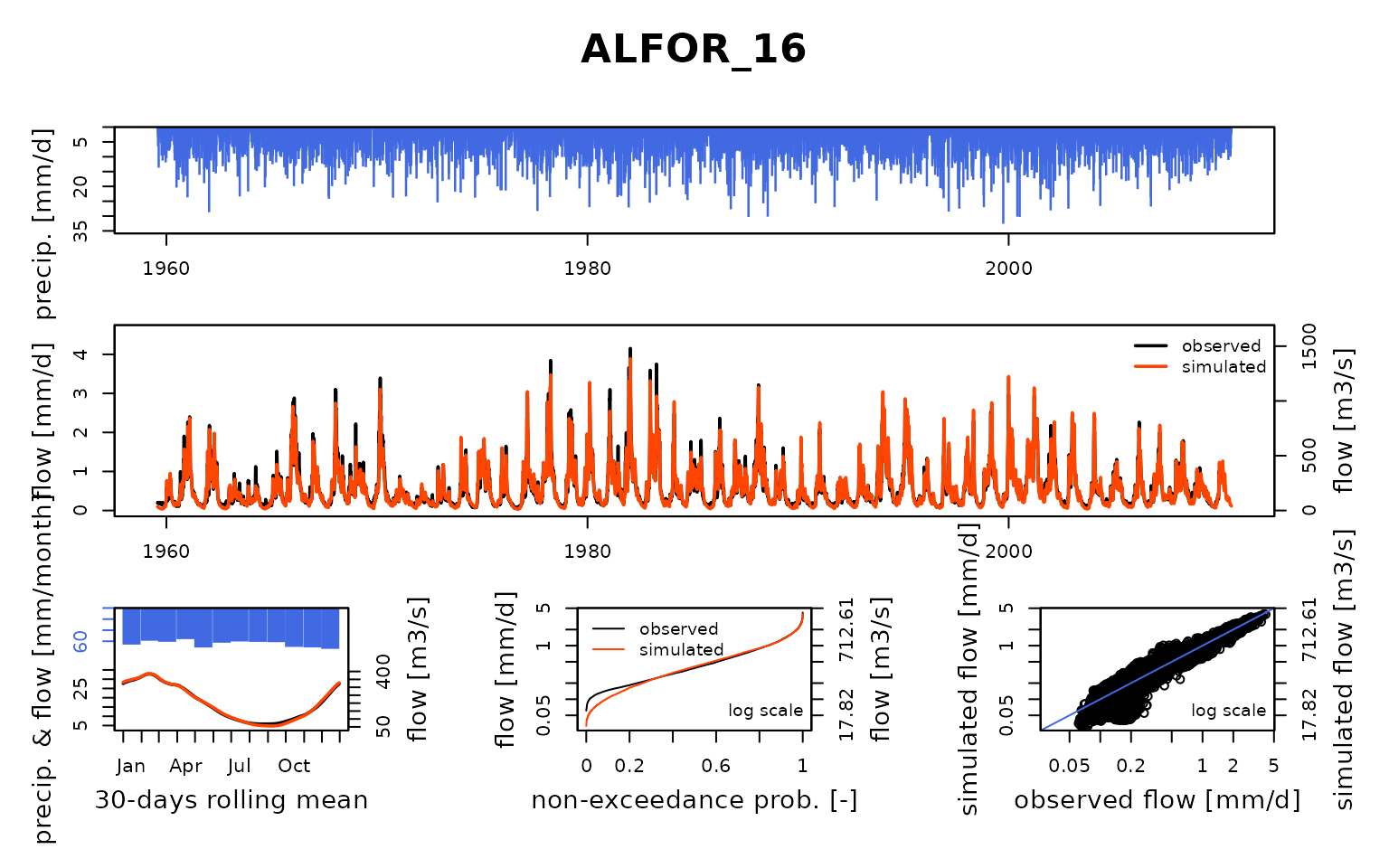
Seine_03: Calibration of a semi-distributed GR model network
David Dorchies
Source:vignettes/seinebasin/V03_First_Calibration.Rmd
V03_First_Calibration.RmdThe aim of this vignette is to perform a first calibration of the semi-distributed hydrological model, using naturalized discharge.
Loading libraries, network and time series data
Run vignette("01_First_network", package = "airGRiwrm")
and vignette("02_First_run", package = "airGRiwrm") before
this one in order to create the Rdata files loaded below:
## Loading required package: airGR##
## Attaching package: 'airGRiwrm'## The following objects are masked from 'package:airGR':
##
## Calibration, CreateCalibOptions, CreateInputsCrit,
## CreateInputsModel, CreateRunOptions, RunModelInputsCrit object
We need then to prepare the InputsCrit object that is necessary to define the calibration objective function. We chose here the KGE’ criterion:
InputsCrit <- CreateInputsCrit(
InputsModel = InputsModel,
FUN_CRIT = ErrorCrit_KGE2,
RunOptions = RunOptions,
Obs = Qnat[IndPeriod_Run,]
)
str(InputsCrit)## List of 25
## $ TRANN_01:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.0943 0.0832 0.1443 0.1387 0.1498 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:2] "Single" "InputsCrit"
## $ STDIZ_04:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.11 0.155 0.409 0.184 0.258 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:2] "Single" "InputsCrit"
## $ BAR-S_06:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.118 0.111 0.111 0.118 0.111 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:2] "Single" "InputsCrit"
## $ CHAUM_07:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.599 0.239 0.239 0.12 0.2 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:2] "Single" "InputsCrit"
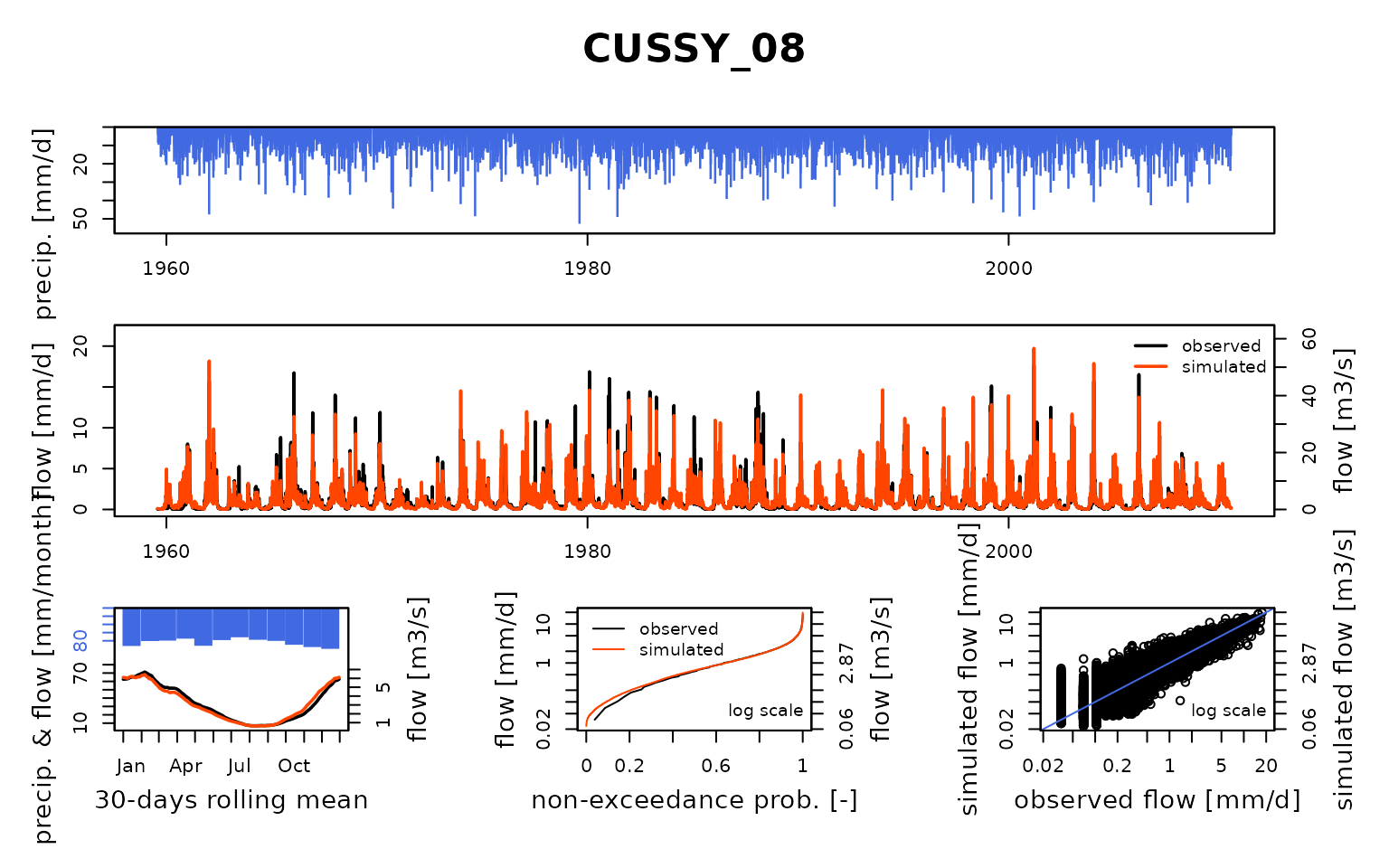
## $ CUSSY_08:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.0697 0.0348 0.0348 0.0348 0.0348 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:2] "Single" "InputsCrit"
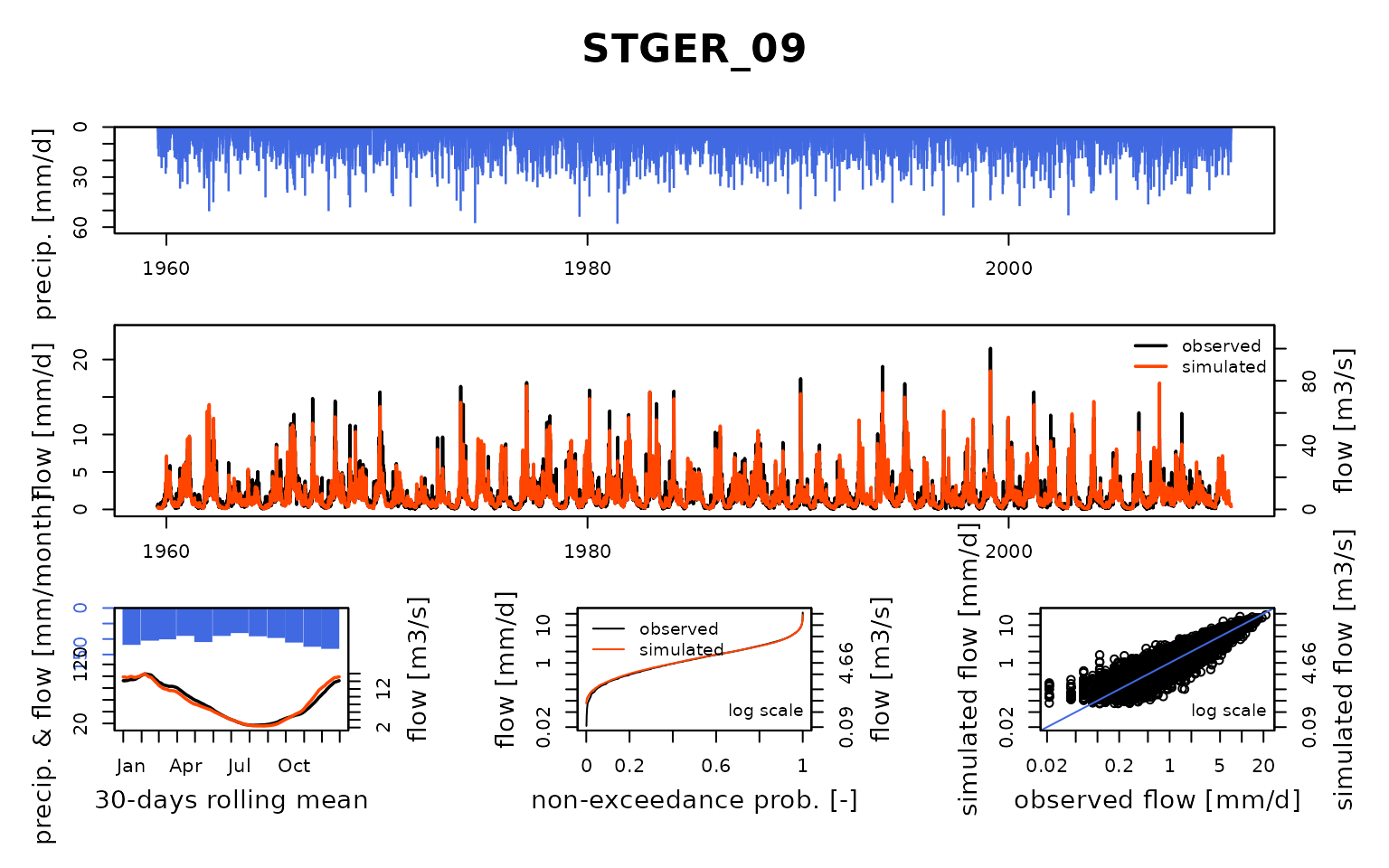
## $ STGER_09:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.579 0.322 0.386 0.429 0.472 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:2] "Single" "InputsCrit"
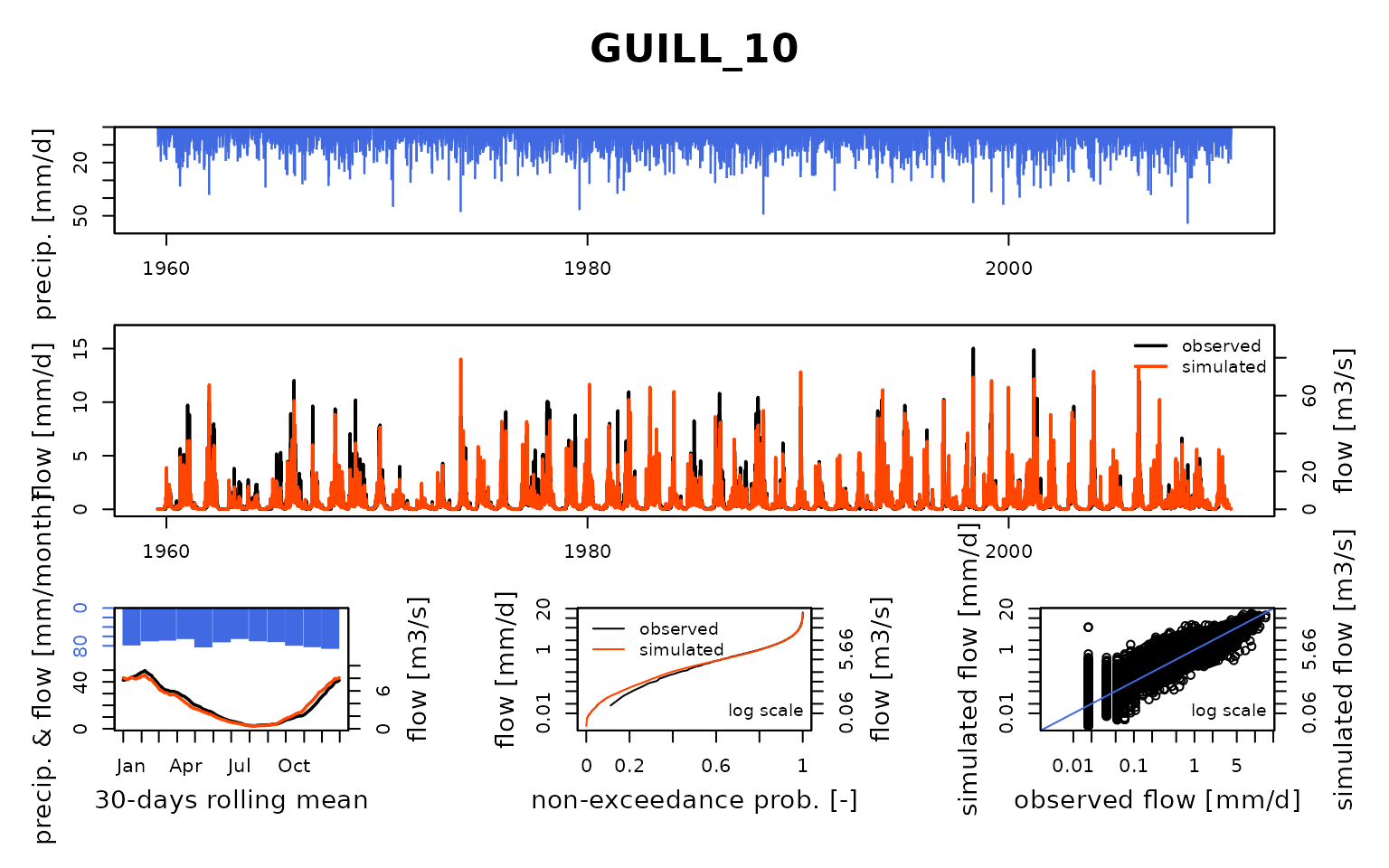
## $ GUILL_10:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.0177 0.0177 0.0177 0.0177 0.0177 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:2] "Single" "InputsCrit"
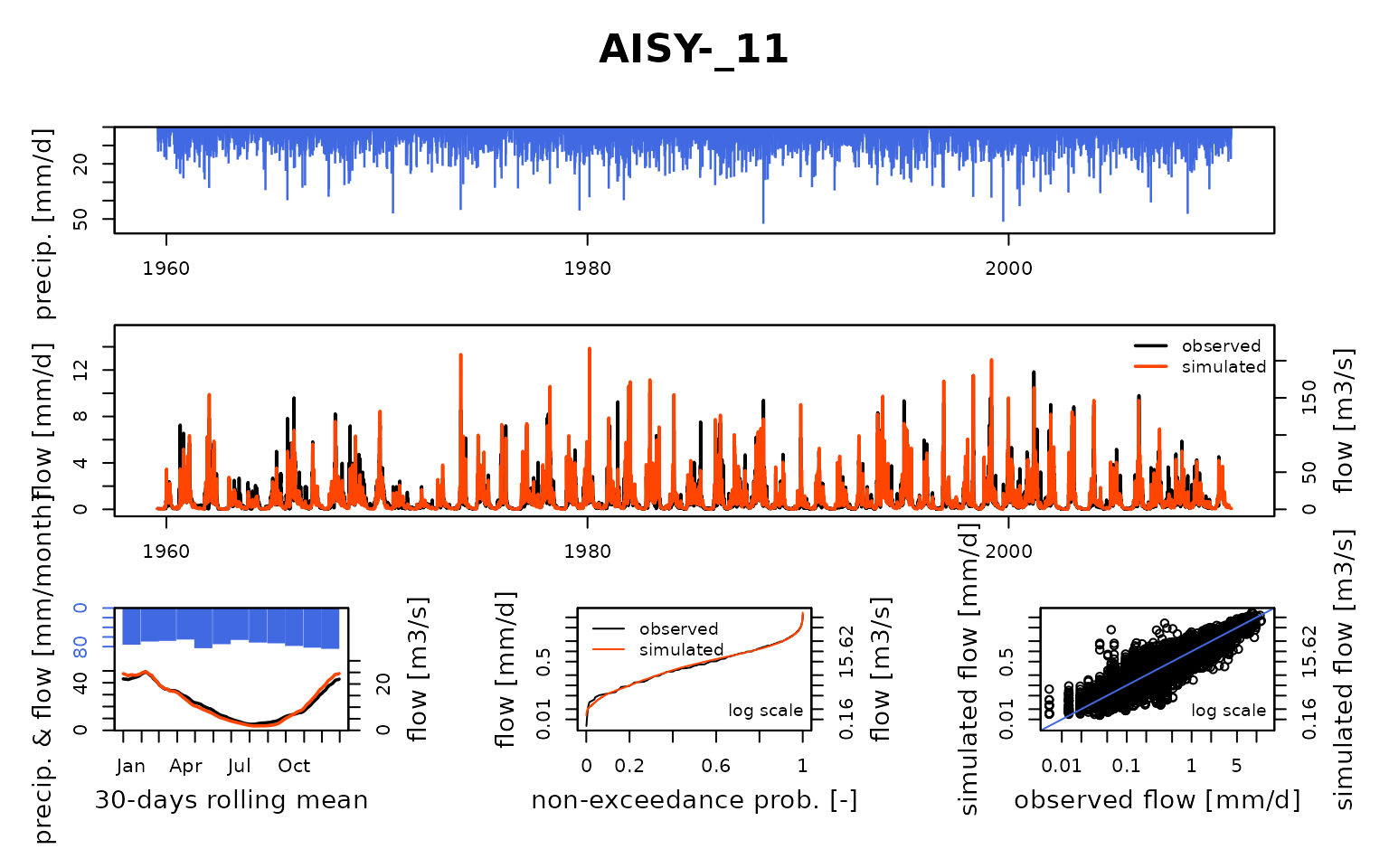
## $ AISY-_11:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.064 0.064 0.064 0.064 0.064 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:2] "Single" "InputsCrit"
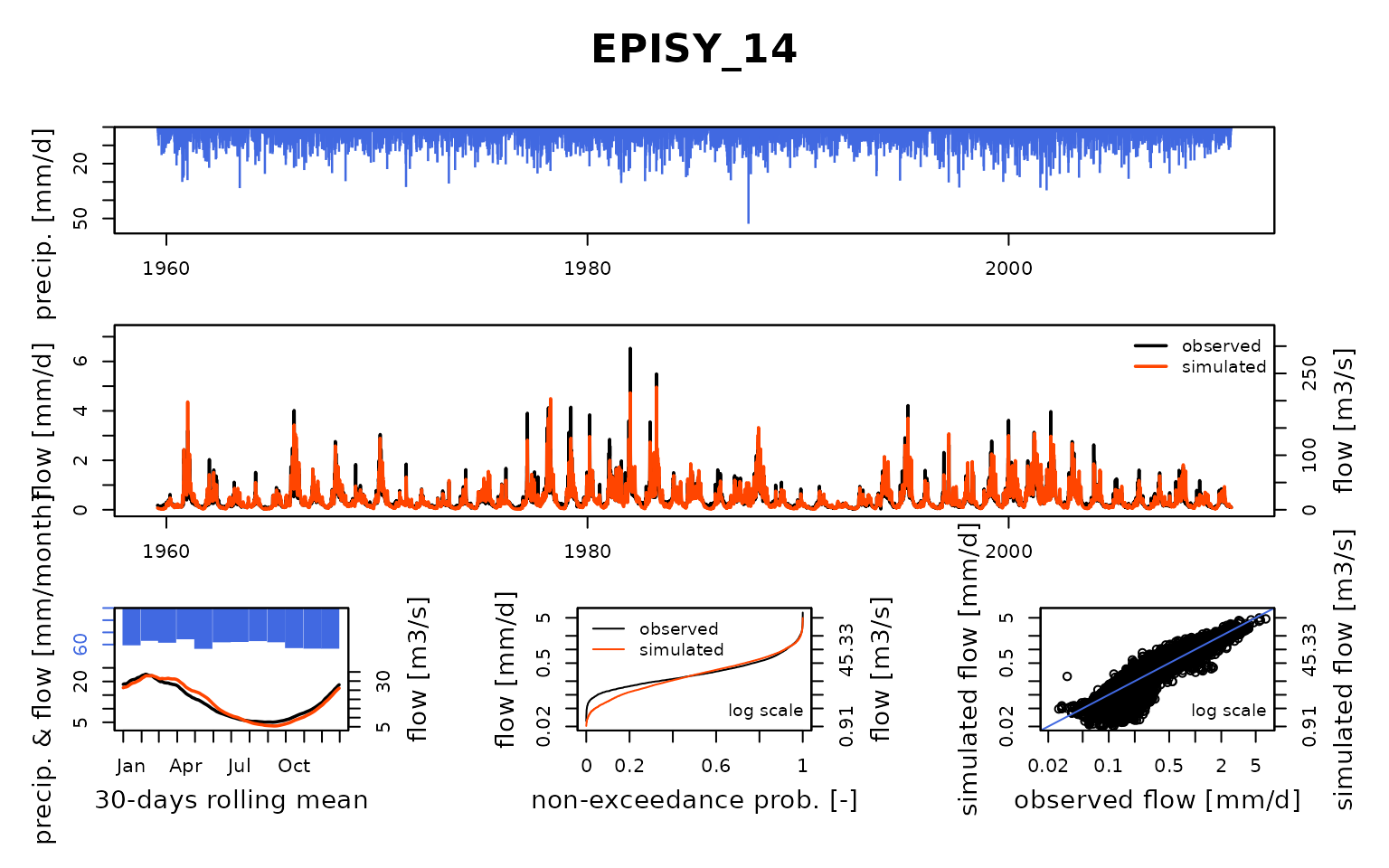
## $ EPISY_14:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.179 0.165 0.159 0.159 0.159 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:2] "Single" "InputsCrit"
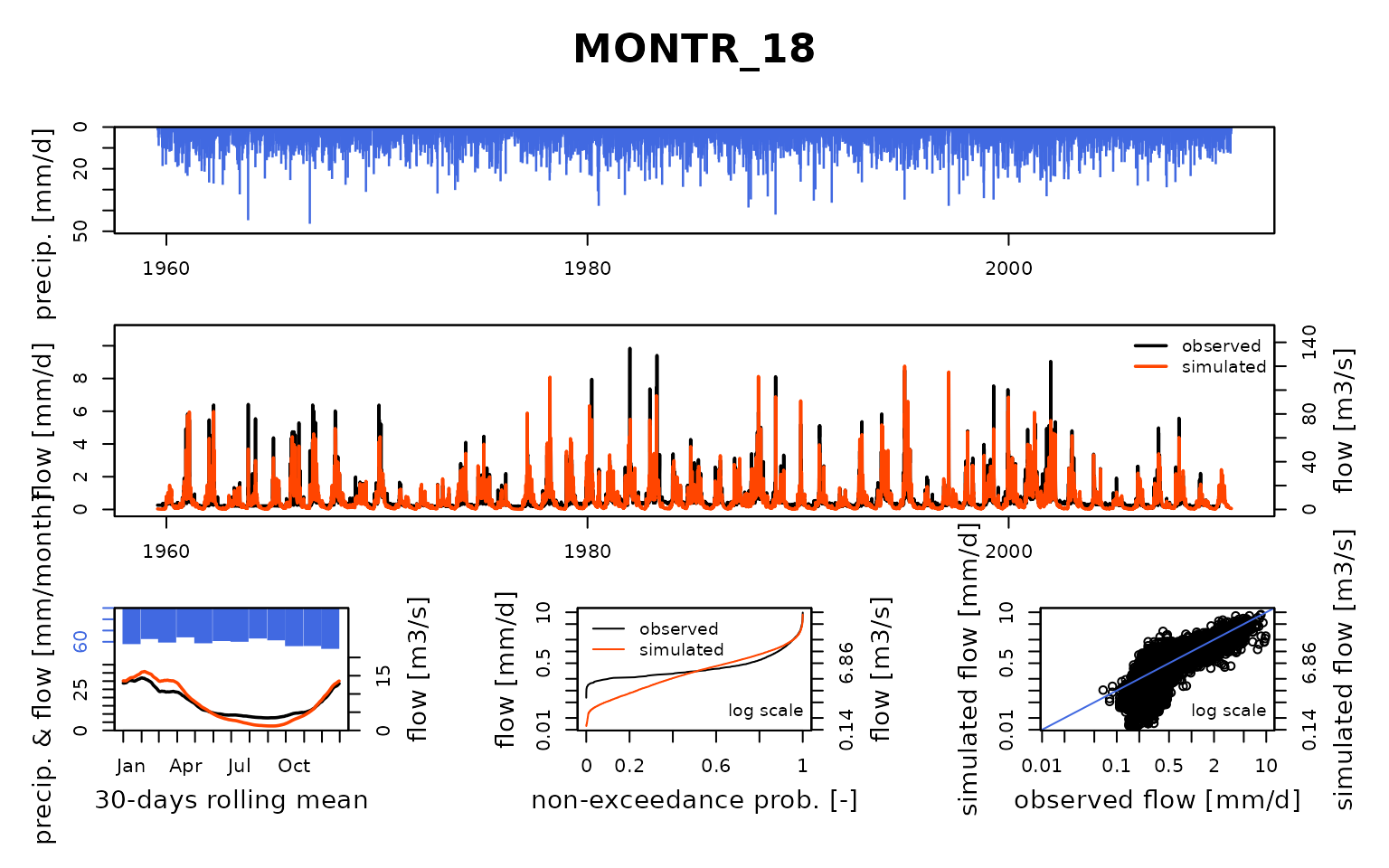
## $ MONTR_18:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.263 0.263 0.263 0.263 0.263 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:2] "Single" "InputsCrit"
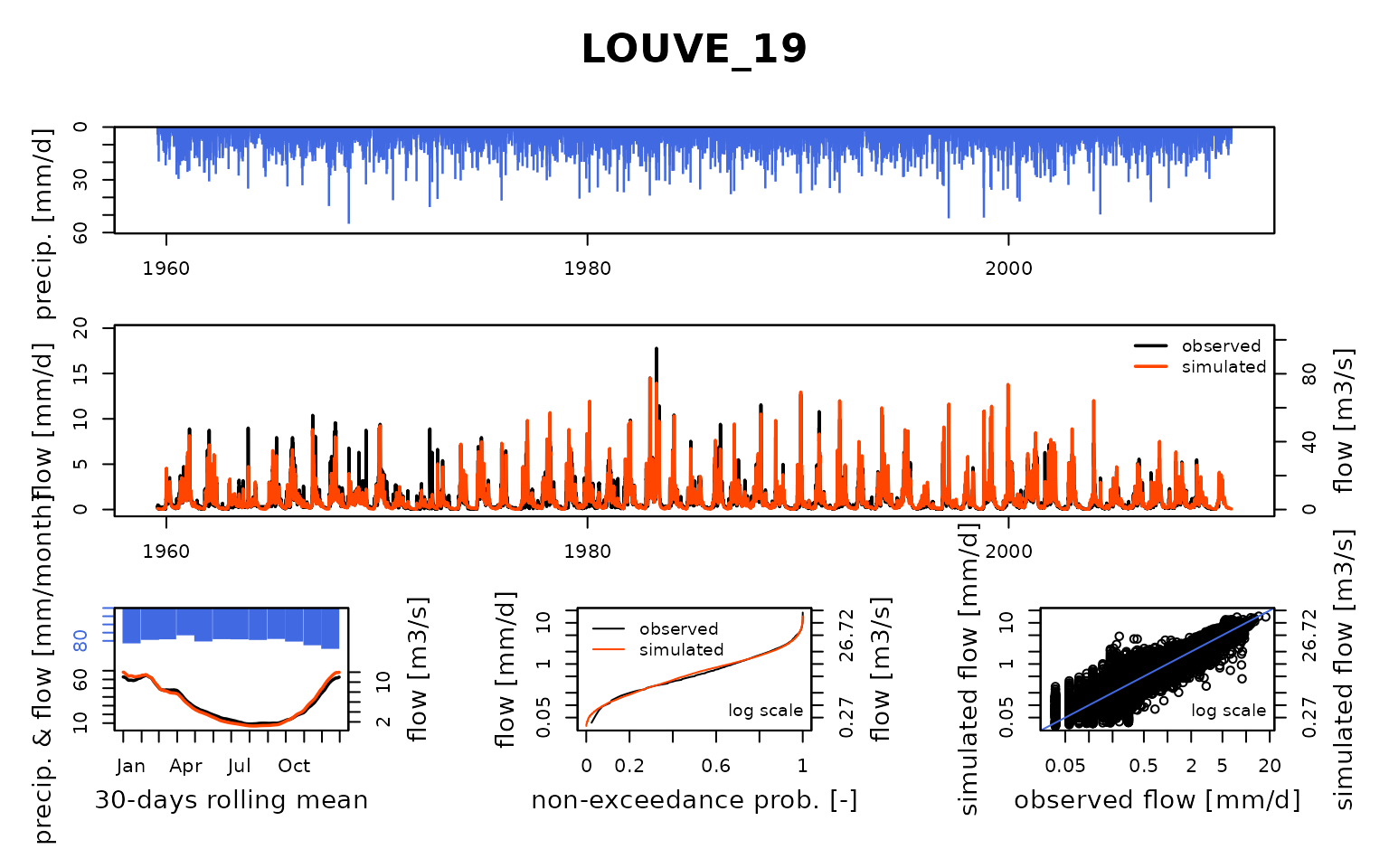
## $ LOUVE_19:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.15 0.187 0.206 0.337 0.281 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:2] "Single" "InputsCrit"
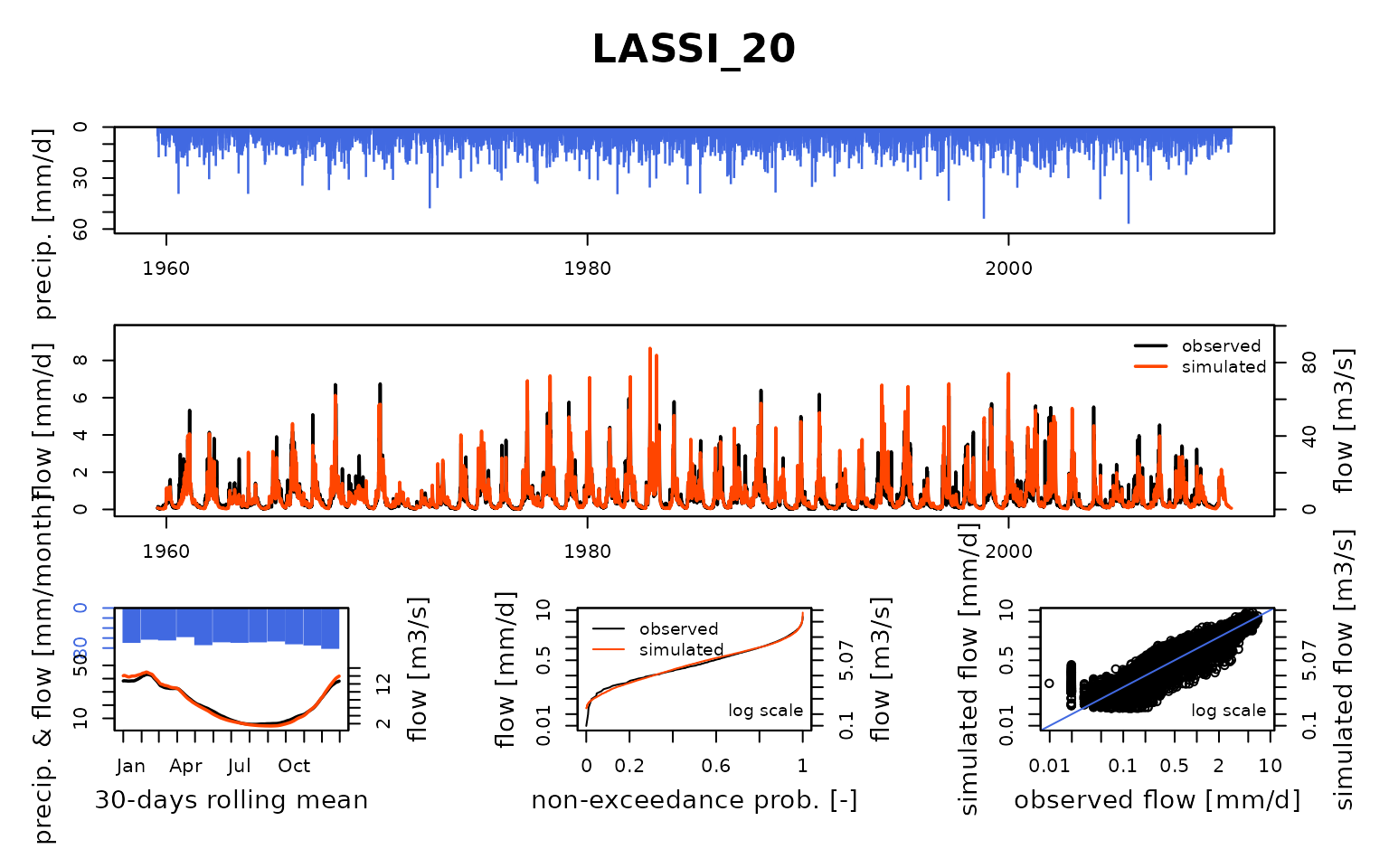
## $ LASSI_20:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.0591 0.0591 0.0986 0.0986 0.0986 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:2] "Single" "InputsCrit"
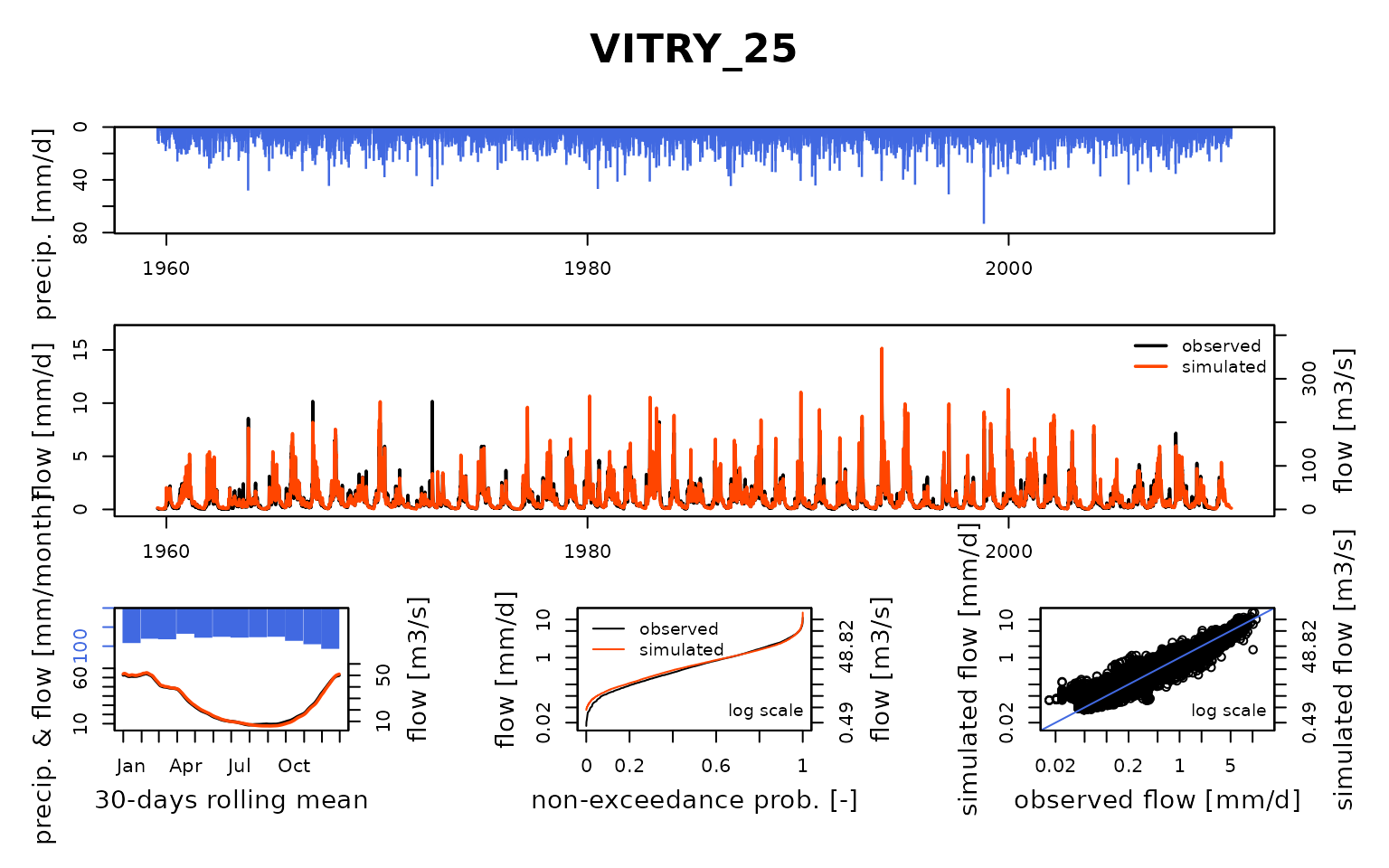
## $ VITRY_25:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.086 0.0737 0.0737 0.0737 0.0737 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:2] "Single" "InputsCrit"
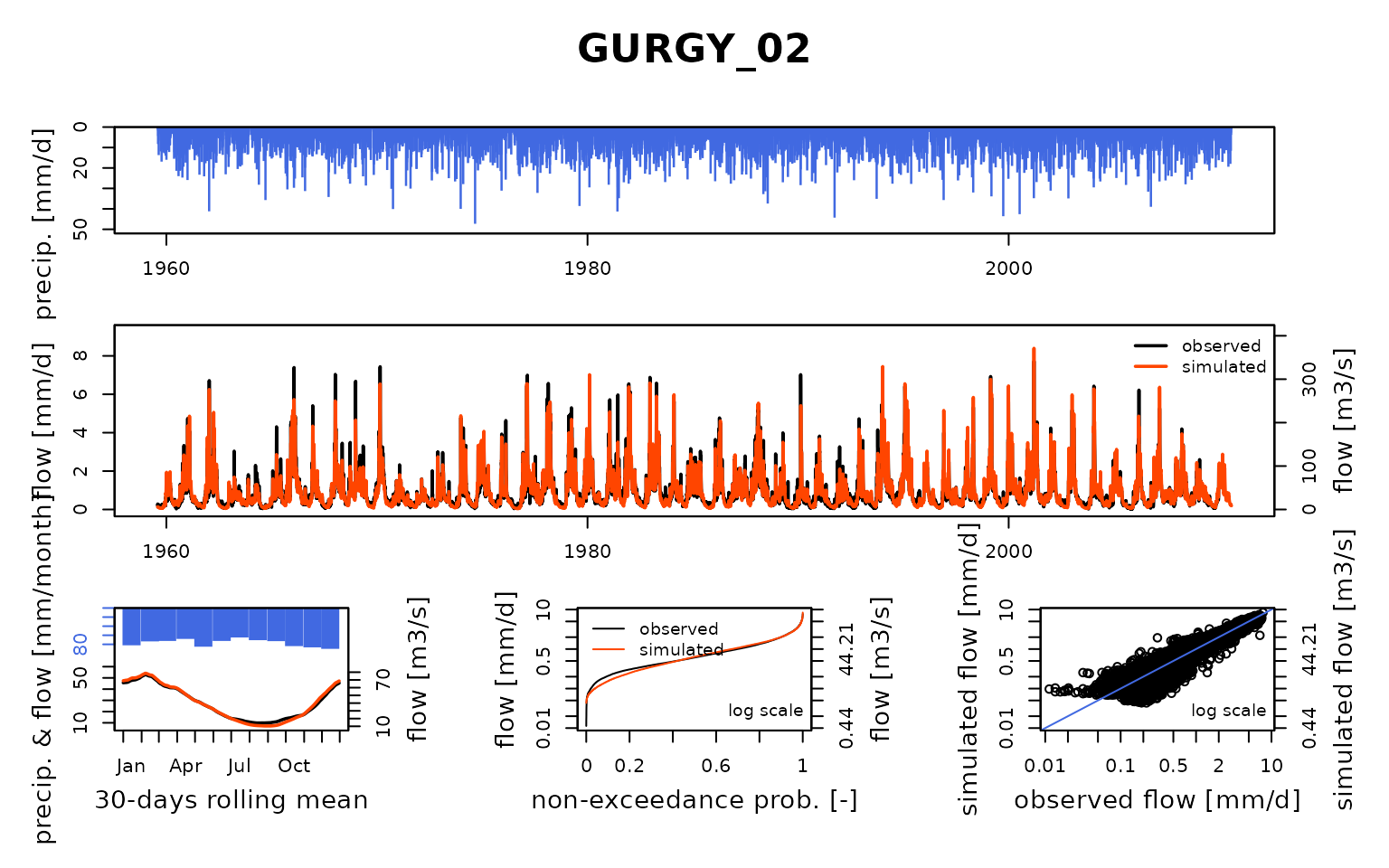
## $ GURGY_02:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.21 0.219 0.285 0.271 0.256 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:3] "InputsCritLavenneFunction" "Single" "InputsCrit"
## ..- attr(*, "Lavenne_FUN")=function (AprParamR, AprCrit)
## ..- attr(*, "AprioriIds")= chr [1:3] "CHAUM_07" "CUSSY_08" "STGER_09"
## ..- attr(*, "AprCelerity")= num 1
## ..- attr(*, "model")=List of 5
## .. ..$ indexParamUngauged: num [1:5] 1 2 3 4 5
## .. ..$ hasX4 : logi TRUE
## .. ..$ iX4 : num 5
## .. ..$ IsHyst : logi FALSE
## .. ..$ X4Ratio : Named num [1:3] 2.19 2.1 1.82
## .. .. ..- attr(*, "names")= chr [1:3] "CHAUM_07" "CUSSY_08" "STGER_09"
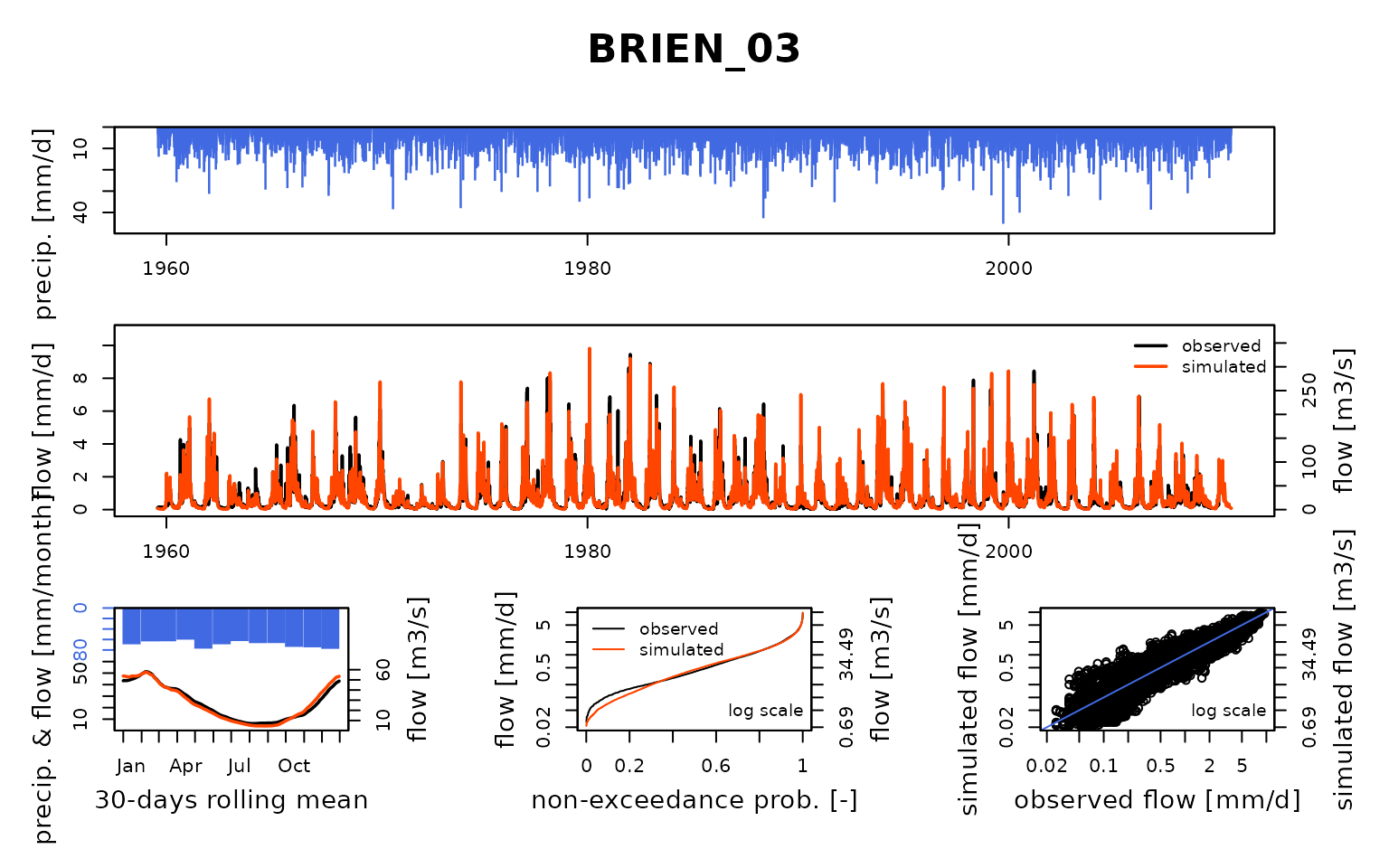
## $ BRIEN_03:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.113 0.104 0.104 0.113 0.104 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:3] "InputsCritLavenneFunction" "Single" "InputsCrit"
## ..- attr(*, "Lavenne_FUN")=function (AprParamR, AprCrit)
## ..- attr(*, "AprioriIds")= chr "AISY-_11"
## ..- attr(*, "AprCelerity")= num 1
## ..- attr(*, "model")=List of 5
## .. ..$ indexParamUngauged: num [1:5] 1 2 3 4 5
## .. ..$ hasX4 : logi TRUE
## .. ..$ iX4 : num 5
## .. ..$ IsHyst : logi FALSE
## .. ..$ X4Ratio : Named num 1.06
## .. .. ..- attr(*, "names")= chr "AISY-_11"
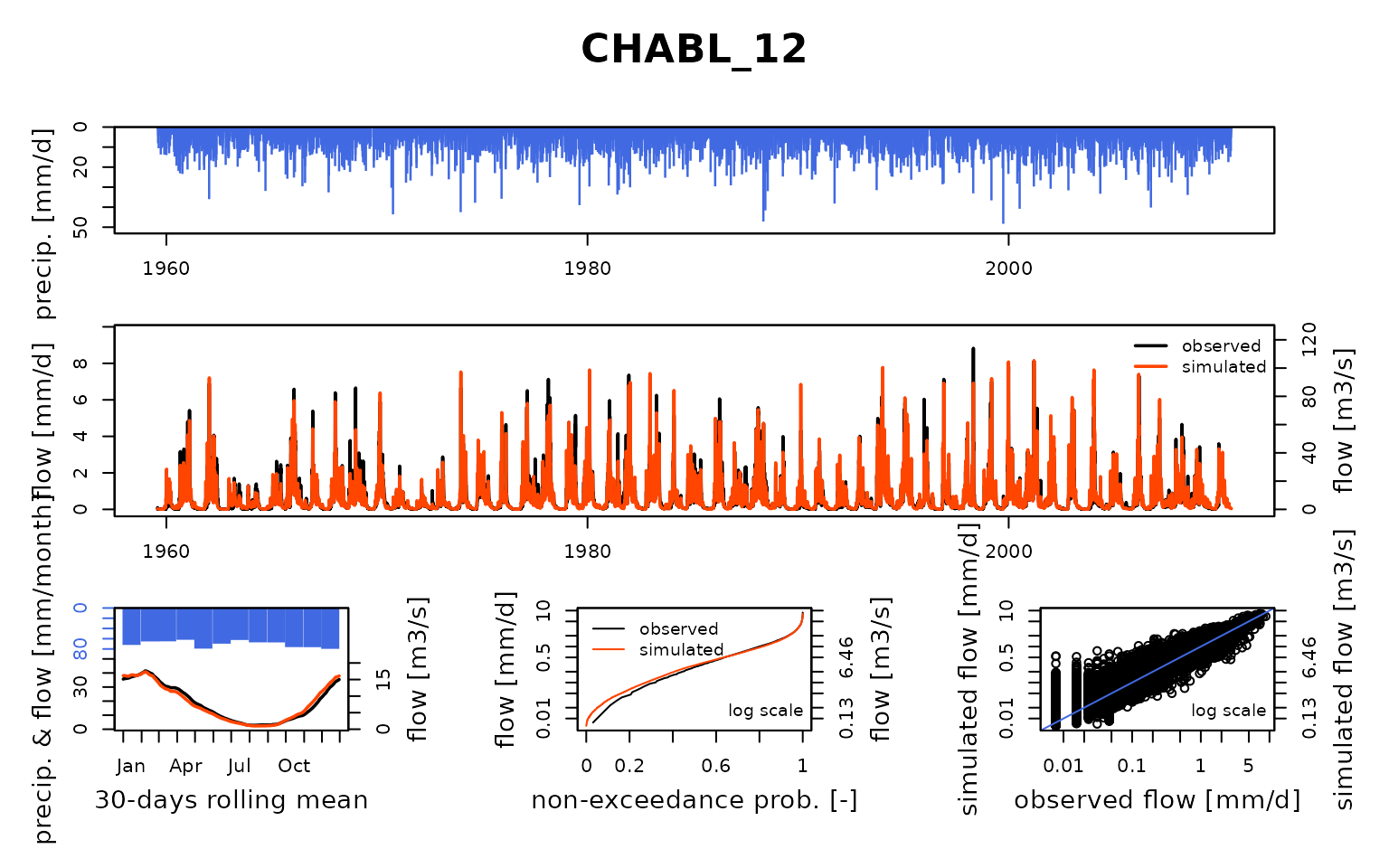
## $ CHABL_12:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.0929 0.0929 0.0155 0.0232 0.0155 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:3] "InputsCritLavenneFunction" "Single" "InputsCrit"
## ..- attr(*, "Lavenne_FUN")=function (AprParamR, AprCrit)
## ..- attr(*, "AprioriIds")= chr "GUILL_10"
## ..- attr(*, "AprCelerity")= num 1
## ..- attr(*, "model")=List of 5
## .. ..$ indexParamUngauged: num [1:5] 1 2 3 4 5
## .. ..$ hasX4 : logi TRUE
## .. ..$ iX4 : num 5
## .. ..$ IsHyst : logi FALSE
## .. ..$ X4Ratio : Named num 1.08
## .. .. ..- attr(*, "names")= chr "GUILL_10"
## $ CHALO_21:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.121 0.157 0.135 0.159 0.264 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:3] "InputsCritLavenneFunction" "Single" "InputsCrit"
## ..- attr(*, "Lavenne_FUN")=function (AprParamR, AprCrit)
## ..- attr(*, "AprioriIds")= chr [1:3] "STDIZ_04" "LOUVE_19" "VITRY_25"
## ..- attr(*, "AprCelerity")= num 1
## ..- attr(*, "model")=List of 5
## .. ..$ indexParamUngauged: num [1:5] 1 2 3 4 5
## .. ..$ hasX4 : logi TRUE
## .. ..$ iX4 : num 5
## .. ..$ IsHyst : logi FALSE
## .. ..$ X4Ratio : Named num [1:3] 0.851 1.387 0.879
## .. .. ..- attr(*, "names")= chr [1:3] "STDIZ_04" "LOUVE_19" "VITRY_25"
## $ MERY-_22:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.0886 0.0975 0.0886 0.0886 0.082 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:3] "InputsCritLavenneFunction" "Single" "InputsCrit"
## ..- attr(*, "Lavenne_FUN")=function (AprParamR, AprCrit)
## ..- attr(*, "AprioriIds")= chr "BAR-S_06"
## ..- attr(*, "AprCelerity")= num 1
## ..- attr(*, "model")=List of 5
## .. ..$ indexParamUngauged: num [1:5] 1 2 3 4 5
## .. ..$ hasX4 : logi TRUE
## .. ..$ iX4 : num 5
## .. ..$ IsHyst : logi FALSE
## .. ..$ X4Ratio : Named num 0.885
## .. .. ..- attr(*, "names")= chr "BAR-S_06"
## $ ARCIS_24:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.0697 0.0529 0.0649 0.0601 0.0961 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:3] "InputsCritLavenneFunction" "Single" "InputsCrit"
## ..- attr(*, "Lavenne_FUN")=function (AprParamR, AprCrit)
## ..- attr(*, "AprioriIds")= chr [1:2] "TRANN_01" "LASSI_20"
## ..- attr(*, "AprCelerity")= num 1
## ..- attr(*, "model")=List of 5
## .. ..$ indexParamUngauged: num [1:5] 1 2 3 4 5
## .. ..$ hasX4 : logi TRUE
## .. ..$ iX4 : num 5
## .. ..$ IsHyst : logi FALSE
## .. ..$ X4Ratio : Named num [1:2] 0.916 1.088
## .. .. ..- attr(*, "names")= chr [1:2] "TRANN_01" "LASSI_20"
## $ NOGEN_13:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.0762 0.0612 0.0612 0.0706 0.0612 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:3] "InputsCritLavenneFunction" "Single" "InputsCrit"
## ..- attr(*, "Lavenne_FUN")=function (AprParamR, AprCrit)
## ..- attr(*, "AprioriIds")= chr [1:2] "MERY-_22" "ARCIS_24"
## ..- attr(*, "AprCelerity")= num 1
## ..- attr(*, "model")=List of 5
## .. ..$ indexParamUngauged: num [1:5] 1 2 3 4 5
## .. ..$ hasX4 : logi TRUE
## .. ..$ iX4 : num 5
## .. ..$ IsHyst : logi FALSE
## .. ..$ X4Ratio : Named num [1:2] 1.02 1.12
## .. .. ..- attr(*, "names")= chr [1:2] "MERY-_22" "ARCIS_24"
## $ NOISI_17:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.139 0.146 0.163 0.17 0.176 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:3] "InputsCritLavenneFunction" "Single" "InputsCrit"
## ..- attr(*, "Lavenne_FUN")=function (AprParamR, AprCrit)
## ..- attr(*, "AprioriIds")= chr [1:2] "MONTR_18" "CHALO_21"
## ..- attr(*, "AprCelerity")= num 1
## ..- attr(*, "model")=List of 5
## .. ..$ indexParamUngauged: num [1:5] 1 2 3 4 5
## .. ..$ hasX4 : logi TRUE
## .. ..$ iX4 : num 5
## .. ..$ IsHyst : logi FALSE
## .. ..$ X4Ratio : Named num [1:2] 1.55 1.48
## .. .. ..- attr(*, "names")= chr [1:2] "MONTR_18" "CHALO_21"
## $ COURL_23:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.205 0.196 0.2 0.218 0.209 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:3] "InputsCritLavenneFunction" "Single" "InputsCrit"
## ..- attr(*, "Lavenne_FUN")=function (AprParamR, AprCrit)
## ..- attr(*, "AprioriIds")= chr [1:3] "GURGY_02" "BRIEN_03" "CHABL_12"
## ..- attr(*, "AprCelerity")= num 1
## ..- attr(*, "model")=List of 5
## .. ..$ indexParamUngauged: num [1:5] 1 2 3 4 5
## .. ..$ hasX4 : logi TRUE
## .. ..$ iX4 : num 5
## .. ..$ IsHyst : logi FALSE
## .. ..$ X4Ratio : Named num [1:3] 0.981 1.173 1.561
## .. .. ..- attr(*, "names")= chr [1:3] "GURGY_02" "BRIEN_03" "CHABL_12"
## $ MONTE_15:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.122 0.129 0.132 0.127 0.137 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:3] "InputsCritLavenneFunction" "Single" "InputsCrit"
## ..- attr(*, "Lavenne_FUN")=function (AprParamR, AprCrit)
## ..- attr(*, "AprioriIds")= chr [1:2] "NOGEN_13" "COURL_23"
## ..- attr(*, "AprCelerity")= num 1
## ..- attr(*, "model")=List of 5
## .. ..$ indexParamUngauged: num [1:5] 1 2 3 4 5
## .. ..$ hasX4 : logi TRUE
## .. ..$ iX4 : num 5
## .. ..$ IsHyst : logi FALSE
## .. ..$ X4Ratio : Named num [1:2] 0.931 0.802
## .. .. ..- attr(*, "names")= chr [1:2] "NOGEN_13" "COURL_23"
## $ ALFOR_16:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.194 0.196 0.198 0.198 0.201 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:3] "InputsCritLavenneFunction" "Single" "InputsCrit"
## ..- attr(*, "Lavenne_FUN")=function (AprParamR, AprCrit)
## ..- attr(*, "AprioriIds")= chr [1:2] "EPISY_14" "MONTE_15"
## ..- attr(*, "AprCelerity")= num 1
## ..- attr(*, "model")=List of 5
## .. ..$ indexParamUngauged: num [1:5] 1 2 3 4 5
## .. ..$ hasX4 : logi TRUE
## .. ..$ iX4 : num 5
## .. ..$ IsHyst : logi FALSE
## .. ..$ X4Ratio : Named num [1:2] 1.12 1.54
## .. .. ..- attr(*, "names")= chr [1:2] "EPISY_14" "MONTE_15"
## $ PARIS_05:List of 8
## ..$ FUN_CRIT:function (InputsCrit, OutputsModel, warnings = TRUE, verbose = TRUE)
## .. ..- attr(*, "class")= chr [1:2] "FUN_CRIT" "function"
## ..$ Obs : num [1:18628] 0.176 0.179 0.183 0.187 0.191 ...
## ..$ VarObs : chr "Q"
## ..$ BoolCrit: logi [1:18628] TRUE TRUE TRUE TRUE TRUE TRUE ...
## ..$ idLayer : logi NA
## ..$ transfo : chr ""
## ..$ epsilon : NULL
## ..$ Weights : NULL
## ..- attr(*, "class")= chr [1:3] "InputsCritLavenneFunction" "Single" "InputsCrit"
## ..- attr(*, "Lavenne_FUN")=function (AprParamR, AprCrit)
## ..- attr(*, "AprioriIds")= chr [1:2] "NOISI_17" "ALFOR_16"
## ..- attr(*, "AprCelerity")= num 1
## ..- attr(*, "model")=List of 5
## .. ..$ indexParamUngauged: num [1:5] 1 2 3 4 5
## .. ..$ hasX4 : logi TRUE
## .. ..$ iX4 : num 5
## .. ..$ IsHyst : logi FALSE
## .. ..$ X4Ratio : Named num [1:2] 0.497 0.48
## .. .. ..- attr(*, "names")= chr [1:2] "NOISI_17" "ALFOR_16"
## - attr(*, "class")= chr [1:2] "GRiwrmInputsCrit" "list"GRiwrmCalibOptions object
CalibOptions <- CreateCalibOptions(InputsModel)
str(CalibOptions)## List of 25
## $ TRANN_01:List of 4
## ..$ FixedParam : logi [1:4] NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:4] 4.59e-05 2.18e+04 -1.09e+04 1.09e+04 4.59e-05 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:4] 169.017 247.151 432.681 -2.376 -0.649 ...
## ..- attr(*, "class")= chr [1:4] "CalibOptions" "daily" "GR" "HBAN"
## $ STDIZ_04:List of 4
## ..$ FixedParam : logi [1:4] NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:4] 4.59e-05 2.18e+04 -1.09e+04 1.09e+04 4.59e-05 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:4] 169.017 247.151 432.681 -2.376 -0.649 ...
## ..- attr(*, "class")= chr [1:4] "CalibOptions" "daily" "GR" "HBAN"
## $ BAR-S_06:List of 4
## ..$ FixedParam : logi [1:4] NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:4] 4.59e-05 2.18e+04 -1.09e+04 1.09e+04 4.59e-05 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:4] 169.017 247.151 432.681 -2.376 -0.649 ...
## ..- attr(*, "class")= chr [1:4] "CalibOptions" "daily" "GR" "HBAN"
## $ CHAUM_07:List of 4
## ..$ FixedParam : logi [1:4] NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:4] 4.59e-05 2.18e+04 -1.09e+04 1.09e+04 4.59e-05 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:4] 169.017 247.151 432.681 -2.376 -0.649 ...
## ..- attr(*, "class")= chr [1:4] "CalibOptions" "daily" "GR" "HBAN"
## $ CUSSY_08:List of 4
## ..$ FixedParam : logi [1:4] NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:4] 4.59e-05 2.18e+04 -1.09e+04 1.09e+04 4.59e-05 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:4] 169.017 247.151 432.681 -2.376 -0.649 ...
## ..- attr(*, "class")= chr [1:4] "CalibOptions" "daily" "GR" "HBAN"
## $ STGER_09:List of 4
## ..$ FixedParam : logi [1:4] NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:4] 4.59e-05 2.18e+04 -1.09e+04 1.09e+04 4.59e-05 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:4] 169.017 247.151 432.681 -2.376 -0.649 ...
## ..- attr(*, "class")= chr [1:4] "CalibOptions" "daily" "GR" "HBAN"
## $ GUILL_10:List of 4
## ..$ FixedParam : logi [1:4] NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:4] 4.59e-05 2.18e+04 -1.09e+04 1.09e+04 4.59e-05 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:4] 169.017 247.151 432.681 -2.376 -0.649 ...
## ..- attr(*, "class")= chr [1:4] "CalibOptions" "daily" "GR" "HBAN"
## $ AISY-_11:List of 4
## ..$ FixedParam : logi [1:4] NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:4] 4.59e-05 2.18e+04 -1.09e+04 1.09e+04 4.59e-05 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:4] 169.017 247.151 432.681 -2.376 -0.649 ...
## ..- attr(*, "class")= chr [1:4] "CalibOptions" "daily" "GR" "HBAN"
## $ EPISY_14:List of 4
## ..$ FixedParam : logi [1:4] NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:4] 4.59e-05 2.18e+04 -1.09e+04 1.09e+04 4.59e-05 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:4] 169.017 247.151 432.681 -2.376 -0.649 ...
## ..- attr(*, "class")= chr [1:4] "CalibOptions" "daily" "GR" "HBAN"
## $ MONTR_18:List of 4
## ..$ FixedParam : logi [1:4] NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:4] 4.59e-05 2.18e+04 -1.09e+04 1.09e+04 4.59e-05 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:4] 169.017 247.151 432.681 -2.376 -0.649 ...
## ..- attr(*, "class")= chr [1:4] "CalibOptions" "daily" "GR" "HBAN"
## $ LOUVE_19:List of 4
## ..$ FixedParam : logi [1:4] NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:4] 4.59e-05 2.18e+04 -1.09e+04 1.09e+04 4.59e-05 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:4] 169.017 247.151 432.681 -2.376 -0.649 ...
## ..- attr(*, "class")= chr [1:4] "CalibOptions" "daily" "GR" "HBAN"
## $ LASSI_20:List of 4
## ..$ FixedParam : logi [1:4] NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:4] 4.59e-05 2.18e+04 -1.09e+04 1.09e+04 4.59e-05 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:4] 169.017 247.151 432.681 -2.376 -0.649 ...
## ..- attr(*, "class")= chr [1:4] "CalibOptions" "daily" "GR" "HBAN"
## $ VITRY_25:List of 4
## ..$ FixedParam : logi [1:4] NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:4] 4.59e-05 2.18e+04 -1.09e+04 1.09e+04 4.59e-05 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:4] 169.017 247.151 432.681 -2.376 -0.649 ...
## ..- attr(*, "class")= chr [1:4] "CalibOptions" "daily" "GR" "HBAN"
## $ GURGY_02:List of 4
## ..$ FixedParam : logi [1:5] NA NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:5] 1.00e-02 2.00e+01 4.59e-05 2.18e+04 -1.09e+04 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:5] 1.25 2.5 5 169.02 247.15 ...
## ..- attr(*, "class")= chr [1:5] "CalibOptions" "daily" "GR" "SD" ...
## $ BRIEN_03:List of 4
## ..$ FixedParam : logi [1:5] NA NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:5] 1.00e-02 2.00e+01 4.59e-05 2.18e+04 -1.09e+04 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:5] 1.25 2.5 5 169.02 247.15 ...
## ..- attr(*, "class")= chr [1:5] "CalibOptions" "daily" "GR" "SD" ...
## $ CHABL_12:List of 4
## ..$ FixedParam : logi [1:5] NA NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:5] 1.00e-02 2.00e+01 4.59e-05 2.18e+04 -1.09e+04 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:5] 1.25 2.5 5 169.02 247.15 ...
## ..- attr(*, "class")= chr [1:5] "CalibOptions" "daily" "GR" "SD" ...
## $ CHALO_21:List of 4
## ..$ FixedParam : logi [1:5] NA NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:5] 1.00e-02 2.00e+01 4.59e-05 2.18e+04 -1.09e+04 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:5] 1.25 2.5 5 169.02 247.15 ...
## ..- attr(*, "class")= chr [1:5] "CalibOptions" "daily" "GR" "SD" ...
## $ MERY-_22:List of 4
## ..$ FixedParam : logi [1:5] NA NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:5] 1.00e-02 2.00e+01 4.59e-05 2.18e+04 -1.09e+04 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:5] 1.25 2.5 5 169.02 247.15 ...
## ..- attr(*, "class")= chr [1:5] "CalibOptions" "daily" "GR" "SD" ...
## $ ARCIS_24:List of 4
## ..$ FixedParam : logi [1:5] NA NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:5] 1.00e-02 2.00e+01 4.59e-05 2.18e+04 -1.09e+04 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:5] 1.25 2.5 5 169.02 247.15 ...
## ..- attr(*, "class")= chr [1:5] "CalibOptions" "daily" "GR" "SD" ...
## $ NOGEN_13:List of 4
## ..$ FixedParam : logi [1:5] NA NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:5] 1.00e-02 2.00e+01 4.59e-05 2.18e+04 -1.09e+04 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:5] 1.25 2.5 5 169.02 247.15 ...
## ..- attr(*, "class")= chr [1:5] "CalibOptions" "daily" "GR" "SD" ...
## $ NOISI_17:List of 4
## ..$ FixedParam : logi [1:5] NA NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:5] 1.00e-02 2.00e+01 4.59e-05 2.18e+04 -1.09e+04 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:5] 1.25 2.5 5 169.02 247.15 ...
## ..- attr(*, "class")= chr [1:5] "CalibOptions" "daily" "GR" "SD" ...
## $ COURL_23:List of 4
## ..$ FixedParam : logi [1:5] NA NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:5] 1.00e-02 2.00e+01 4.59e-05 2.18e+04 -1.09e+04 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:5] 1.25 2.5 5 169.02 247.15 ...
## ..- attr(*, "class")= chr [1:5] "CalibOptions" "daily" "GR" "SD" ...
## $ MONTE_15:List of 4
## ..$ FixedParam : logi [1:5] NA NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:5] 1.00e-02 2.00e+01 4.59e-05 2.18e+04 -1.09e+04 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:5] 1.25 2.5 5 169.02 247.15 ...
## ..- attr(*, "class")= chr [1:5] "CalibOptions" "daily" "GR" "SD" ...
## $ ALFOR_16:List of 4
## ..$ FixedParam : logi [1:5] NA NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:5] 1.00e-02 2.00e+01 4.59e-05 2.18e+04 -1.09e+04 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:5] 1.25 2.5 5 169.02 247.15 ...
## ..- attr(*, "class")= chr [1:5] "CalibOptions" "daily" "GR" "SD" ...
## $ PARIS_05:List of 4
## ..$ FixedParam : logi [1:5] NA NA NA NA NA
## ..$ SearchRanges : num [1:2, 1:5] 1.00e-02 2.00e+01 4.59e-05 2.18e+04 -1.09e+04 ...
## ..$ FUN_TRANSFO :function (ParamIn, Direction)
## ..$ StartParamDistrib: num [1:3, 1:5] 1.25 2.5 5 169.02 247.15 ...
## ..- attr(*, "class")= chr [1:5] "CalibOptions" "daily" "GR" "SD" ...
## - attr(*, "class")= chr [1:2] "GRiwrmCalibOptions" "list"Calibration
The optimization (i.e. calibration) of parameters can now be performed:
OutputsCalib <- Calibration(InputsModel, RunOptions, InputsCrit, CalibOptions)## Calibration.GRiwrmInputsModel: Processing sub-basin 'TRANN_01'...## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (81 runs)
## Param = 247.151, -0.020, 83.096, 2.384
## Crit. KGE2[Q] = 0.8828
## Steepest-descent local search in progress
## Calibration completed (47 iterations, 454 runs)
## Param = 202.172, -0.124, 76.792, 5.505
## Crit. KGE2[Q] = 0.9549
## Calibration.GRiwrmInputsModel: Processing sub-basin 'STDIZ_04'...
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (81 runs)
## Param = 169.017, -0.020, 83.096, 2.384
## Crit. KGE2[Q] = 0.9015
## Steepest-descent local search in progress
## Calibration completed (34 iterations, 342 runs)
## Param = 161.253, -0.238, 69.848, 3.746
## Crit. KGE2[Q] = 0.9472
## Calibration.GRiwrmInputsModel: Processing sub-basin 'BAR-S_06'...
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (81 runs)
## Param = 247.151, -0.020, 83.096, 2.384
## Crit. KGE2[Q] = 0.8826
## Steepest-descent local search in progress
## Calibration completed (27 iterations, 281 runs)
## Param = 247.151, -0.347, 91.836, 5.302
## Crit. KGE2[Q] = 0.9588
## Calibration.GRiwrmInputsModel: Processing sub-basin 'CHAUM_07'...
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (81 runs)
## Param = 432.681, -0.020, 83.096, 1.417
## Crit. KGE2[Q] = 0.9019
## Steepest-descent local search in progress
## Calibration completed (32 iterations, 331 runs)
## Param = 269.272, 0.421, 166.665, 1.257
## Crit. KGE2[Q] = 0.9434
## Calibration.GRiwrmInputsModel: Processing sub-basin 'CUSSY_08'...
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (81 runs)
## Param = 247.151, -0.649, 42.098, 2.384
## Crit. KGE2[Q] = 0.8985
## Steepest-descent local search in progress
## Calibration completed (26 iterations, 278 runs)
## Param = 214.586, -0.974, 59.761, 2.087
## Crit. KGE2[Q] = 0.9066
## Calibration.GRiwrmInputsModel: Processing sub-basin 'STGER_09'...
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (81 runs)
## Param = 432.681, -0.020, 83.096, 1.417
## Crit. KGE2[Q] = 0.9225
## Steepest-descent local search in progress
## Calibration completed (28 iterations, 297 runs)
## Param = 325.478, -0.313, 142.419, 1.145
## Crit. KGE2[Q] = 0.9376
## Calibration.GRiwrmInputsModel: Processing sub-basin 'GUILL_10'...
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (81 runs)
## Param = 169.017, -2.376, 42.098, 2.384
## Crit. KGE2[Q] = 0.8562
## Steepest-descent local search in progress
## Calibration completed (32 iterations, 331 runs)
## Param = 183.842, -1.733, 24.004, 2.586
## Crit. KGE2[Q] = 0.8879
## Calibration.GRiwrmInputsModel: Processing sub-basin 'AISY-_11'...
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (81 runs)
## Param = 169.017, -2.376, 83.096, 2.384
## Crit. KGE2[Q] = 0.8725
## Steepest-descent local search in progress
## Calibration completed (41 iterations, 411 runs)
## Param = 172.375, -1.653, 49.462, 2.668
## Crit. KGE2[Q] = 0.9175
## Calibration.GRiwrmInputsModel: Processing sub-basin 'EPISY_14'...
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (81 runs)
## Param = 432.681, -0.649, 83.096, 2.384
## Crit. KGE2[Q] = 0.8520
## Steepest-descent local search in progress
## Calibration completed (36 iterations, 369 runs)
## Param = 610.285, -0.593, 41.294, 3.754
## Crit. KGE2[Q] = 0.9395
## Calibration.GRiwrmInputsModel: Processing sub-basin 'MONTR_18'...
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (81 runs)
## Param = 247.151, -0.649, 42.098, 2.384
## Crit. KGE2[Q] = 0.8528
## Steepest-descent local search in progress
## Calibration completed (18 iterations, 208 runs)
## Param = 292.949, -0.578, 40.854, 2.169
## Crit. KGE2[Q] = 0.8634
## Calibration.GRiwrmInputsModel: Processing sub-basin 'LOUVE_19'...
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (81 runs)
## Param = 169.017, -0.649, 83.096, 2.384
## Crit. KGE2[Q] = 0.8814
## Steepest-descent local search in progress
## Calibration completed (17 iterations, 200 runs)
## Param = 162.390, -1.099, 83.096, 3.438
## Crit. KGE2[Q] = 0.9138
## Calibration.GRiwrmInputsModel: Processing sub-basin 'LASSI_20'...
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (81 runs)
## Param = 247.151, -0.649, 83.096, 2.384
## Crit. KGE2[Q] = 0.8510
## Steepest-descent local search in progress
## Calibration completed (32 iterations, 322 runs)
## Param = 223.632, -1.160, 75.944, 4.355
## Crit. KGE2[Q] = 0.9075
## Calibration.GRiwrmInputsModel: Processing sub-basin 'VITRY_25'...
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (81 runs)
## Param = 432.681, -0.649, 83.096, 2.384
## Crit. KGE2[Q] = 0.8652
## Steepest-descent local search in progress
## Calibration completed (83 iterations, 785 runs)
## Param = 278.332, -1.382, 97.715, 5.076
## Crit. KGE2[Q] = 0.9550
## Calibration.GRiwrmInputsModel: Processing sub-basin 'GURGY_02'...
## Parameter regularization: test a priori parameters from node CHAUM_07: 1, 269.272, 0.421, 166.665, 2.753
## Crit. KGE2[Q] = 0.6520
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9585
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 0.9734
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.3445
##
## Parameter regularization: test a priori parameters from node CUSSY_08: 1, 214.586, -0.974, 59.761, 4.388
## Crit. KGE2[Q] = 0.7694
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9359
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.1511
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.1620
##
## Parameter regularization: test a priori parameters from node STGER_09: 1, 325.478, -0.313, 142.419, 2.082
## Crit. KGE2[Q] = 0.7499
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9586
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 0.9852
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.2462
##
## Parameter regularization: set a priori parameters from node CUSSY_08: 1, 214.586, -0.974, 59.761, 4.388
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (243 runs)
## Param = 1.250, 432.681, -2.376, 83.096, 2.384
## Crit. Composite = 0.9123
## Steepest-descent local search in progress
## Calibration completed (60 iterations, 845 runs)
## Param = 1.422, 520.044, -2.610, 100.230, 4.052
## Crit. Composite = 0.9507
## Formula: sum(0.88 * KGE2[sqrt(Q)], 0.12 * GAPX[ParamT])
## Calibration.GRiwrmInputsModel: Processing sub-basin 'BRIEN_03'...
## Parameter regularization: get a priori parameters from node AISY-_11: 1, 172.375, -1.653, 49.462, 2.824
## Crit. KGE2[Q] = 0.8457
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9178
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.0549
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 0.8816
##
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (243 runs)
## Param = 1.250, 247.151, -0.649, 83.096, 2.384
## Crit. Composite = 0.9456
## Steepest-descent local search in progress
## Calibration completed (24 iterations, 460 runs)
## Param = 0.540, 219.203, -0.432, 81.451, 2.813
## Crit. Composite = 0.9517
## Formula: sum(0.87 * KGE2[sqrt(Q)], 0.13 * GAPX[ParamT])
## Calibration.GRiwrmInputsModel: Processing sub-basin 'CHABL_12'...
## Parameter regularization: get a priori parameters from node GUILL_10: 1, 183.842, -1.733, 24.004, 2.788
## Crit. KGE2[Q] = 0.8336
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.8707
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.1026
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.0215
##
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (243 runs)
## Param = 1.250, 169.017, -2.376, 42.098, 2.384
## Crit. Composite = 0.9191
## Steepest-descent local search in progress
## Calibration completed (31 iterations, 531 runs)
## Param = 0.318, 195.523, -2.557, 34.220, 3.050
## Crit. Composite = 0.9380
## Formula: sum(0.87 * KGE2[sqrt(Q)], 0.13 * GAPX[ParamT])
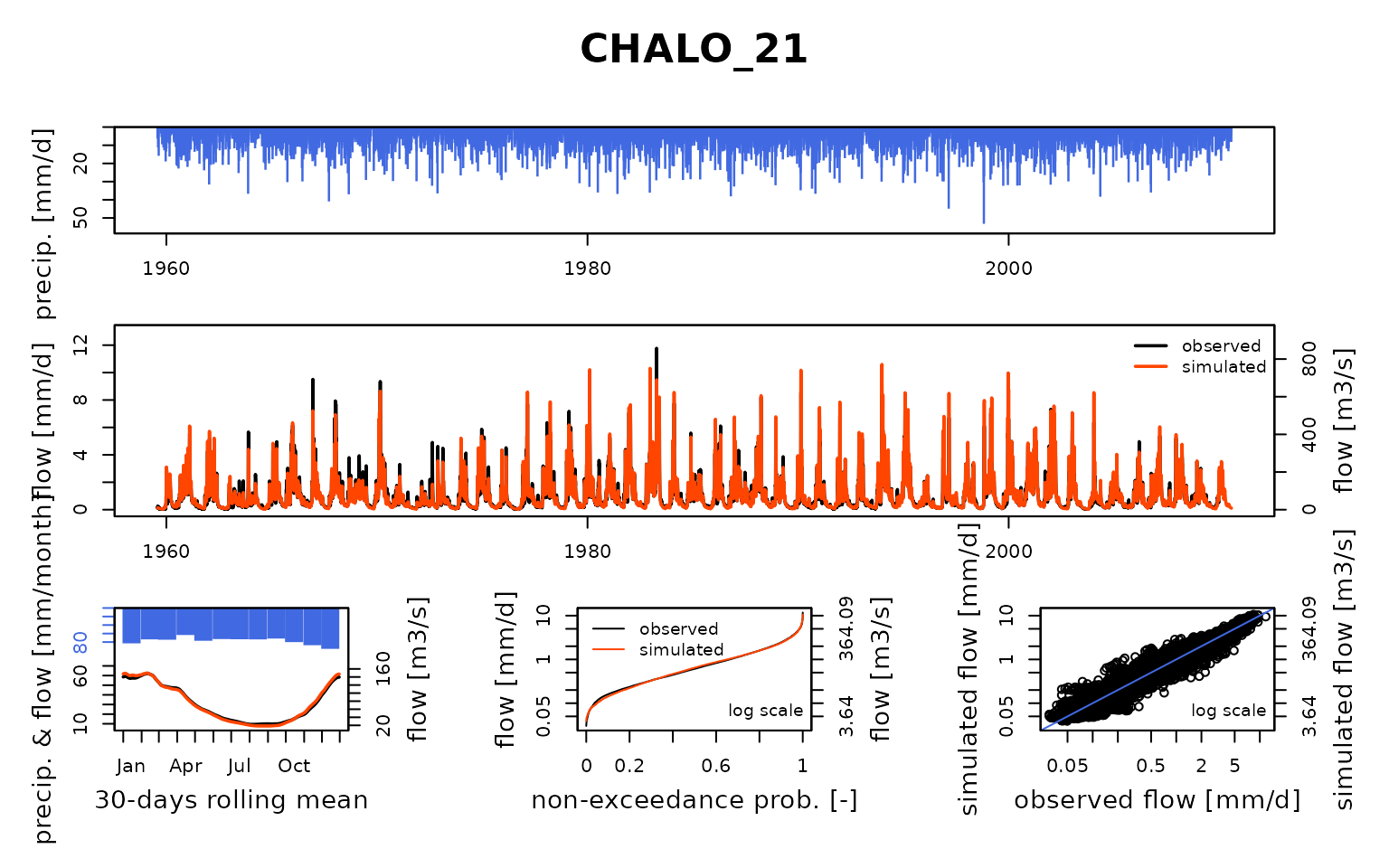
## Calibration.GRiwrmInputsModel: Processing sub-basin 'CHALO_21'...
## Parameter regularization: test a priori parameters from node STDIZ_04: 1, 161.253, -0.238, 69.848, 3.19
## Crit. KGE2[Q] = 0.8681
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9246
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.0701
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.0825
##
## Parameter regularization: test a priori parameters from node LOUVE_19: 1, 162.39, -1.099, 83.096, 4.767
## Crit. KGE2[Q] = 0.8834
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9356
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.0807
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.0541
##
## Parameter regularization: test a priori parameters from node VITRY_25: 1, 278.332, -1.382, 97.715, 4.463
## Crit. KGE2[Q] = 0.9105
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9406
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.0551
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.0380
##
## Parameter regularization: set a priori parameters from node VITRY_25: 1, 278.332, -1.382, 97.715, 4.463
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (243 runs)
## Param = 1.250, 432.681, -0.649, 83.096, 2.384
## Crit. Composite = 0.9372
## Steepest-descent local search in progress
## Calibration completed (37 iterations, 591 runs)
## Param = 0.420, 601.845, -2.060, 113.296, 4.423
## Crit. Composite = 0.9658
## Formula: sum(0.86 * KGE2[sqrt(Q)], 0.14 * GAPX[ParamT])
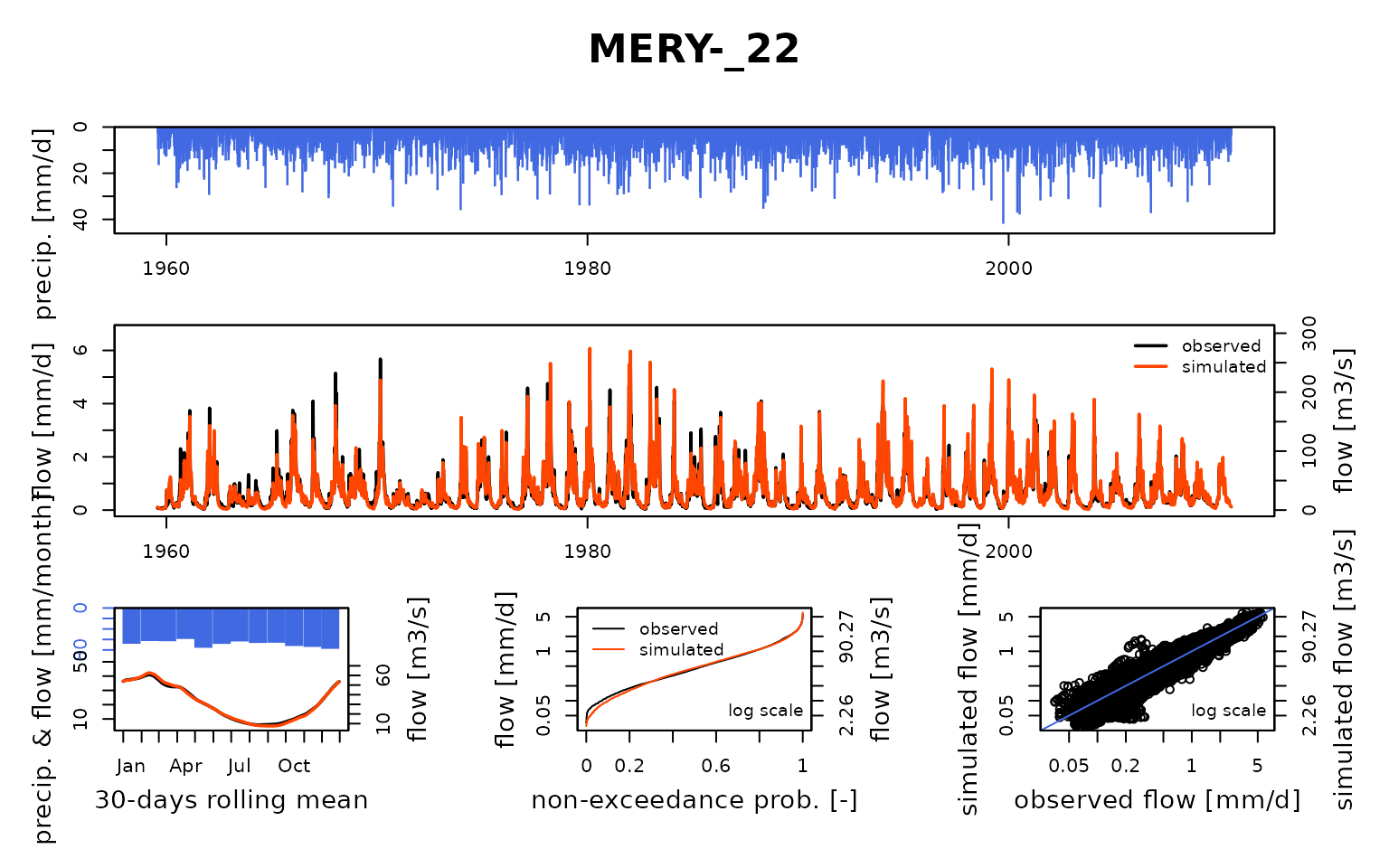
## Calibration.GRiwrmInputsModel: Processing sub-basin 'MERY-_22'...
## Parameter regularization: get a priori parameters from node BAR-S_06: 1, 247.151, -0.347, 91.836, 4.694
## Crit. KGE2[Q] = 0.7652
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9287
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.0475
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.2186
##
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (243 runs)
## Param = 1.250, 432.681, -2.376, 83.096, 2.384
## Crit. Composite = 0.9225
## Steepest-descent local search in progress
## Calibration completed (40 iterations, 621 runs)
## Param = 0.330, 749.945, -3.589, 60.340, 4.638
## Crit. Composite = 0.9523
## Formula: sum(0.88 * KGE2[sqrt(Q)], 0.12 * GAPX[ParamT])
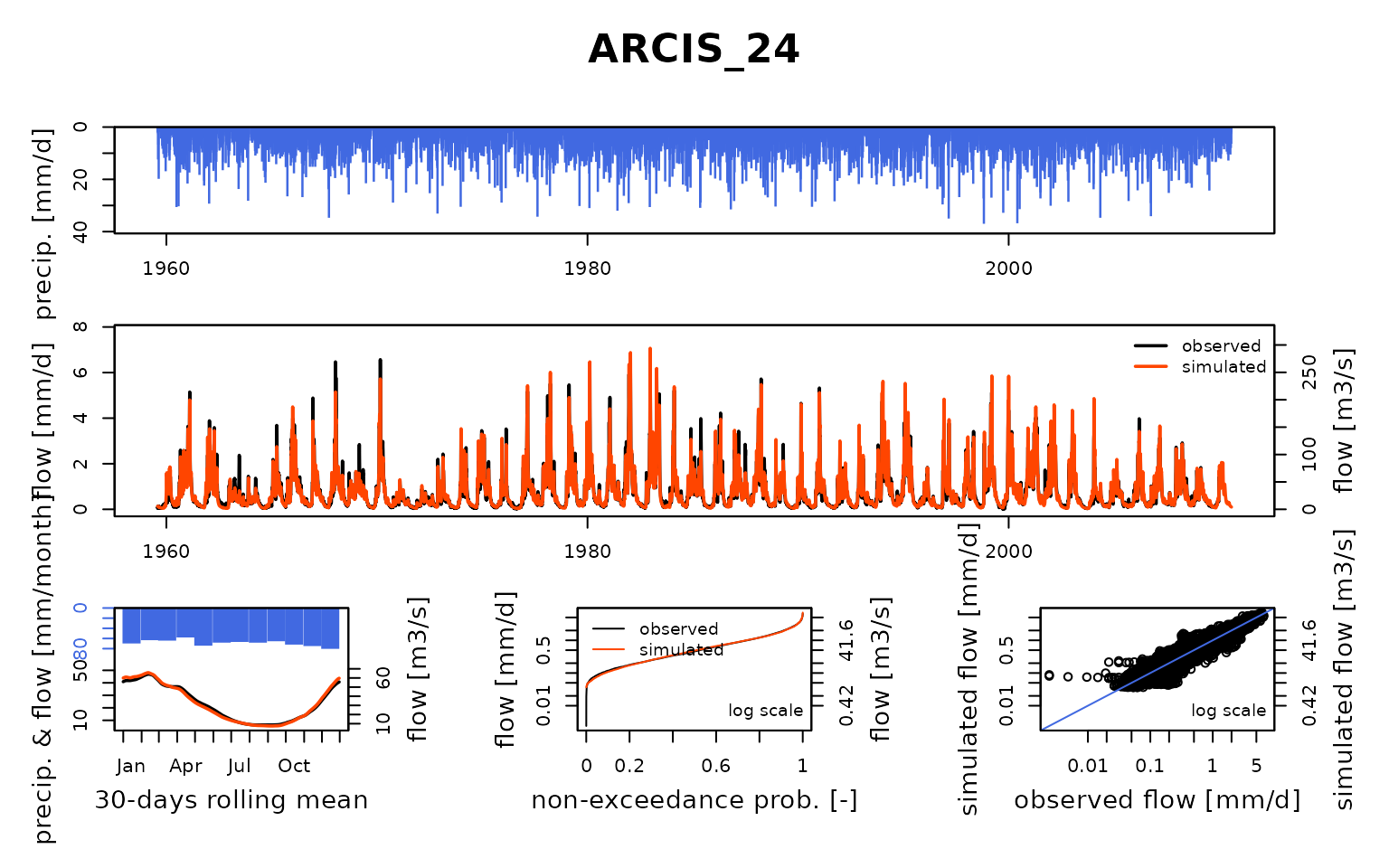
## Calibration.GRiwrmInputsModel: Processing sub-basin 'ARCIS_24'...
## Parameter regularization: test a priori parameters from node TRANN_01: 1, 202.172, -0.124, 76.792, 5.041
## Crit. KGE2[Q] = 0.8186
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9318
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.0529
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.1596
##
## Parameter regularization: test a priori parameters from node LASSI_20: 1, 223.632, -1.16, 75.944, 4.738
## Crit. KGE2[Q] = 0.8700
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9328
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.0574
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.0954
##
## Parameter regularization: set a priori parameters from node LASSI_20: 1, 223.632, -1.16, 75.944, 4.738
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (243 runs)
## Param = 1.250, 432.681, -2.376, 83.096, 2.384
## Crit. Composite = 0.9379
## Steepest-descent local search in progress
## Calibration completed (30 iterations, 520 runs)
## Param = 0.320, 539.153, -2.973, 81.451, 4.687
## Crit. Composite = 0.9600
## Formula: sum(0.87 * KGE2[sqrt(Q)], 0.13 * GAPX[ParamT])
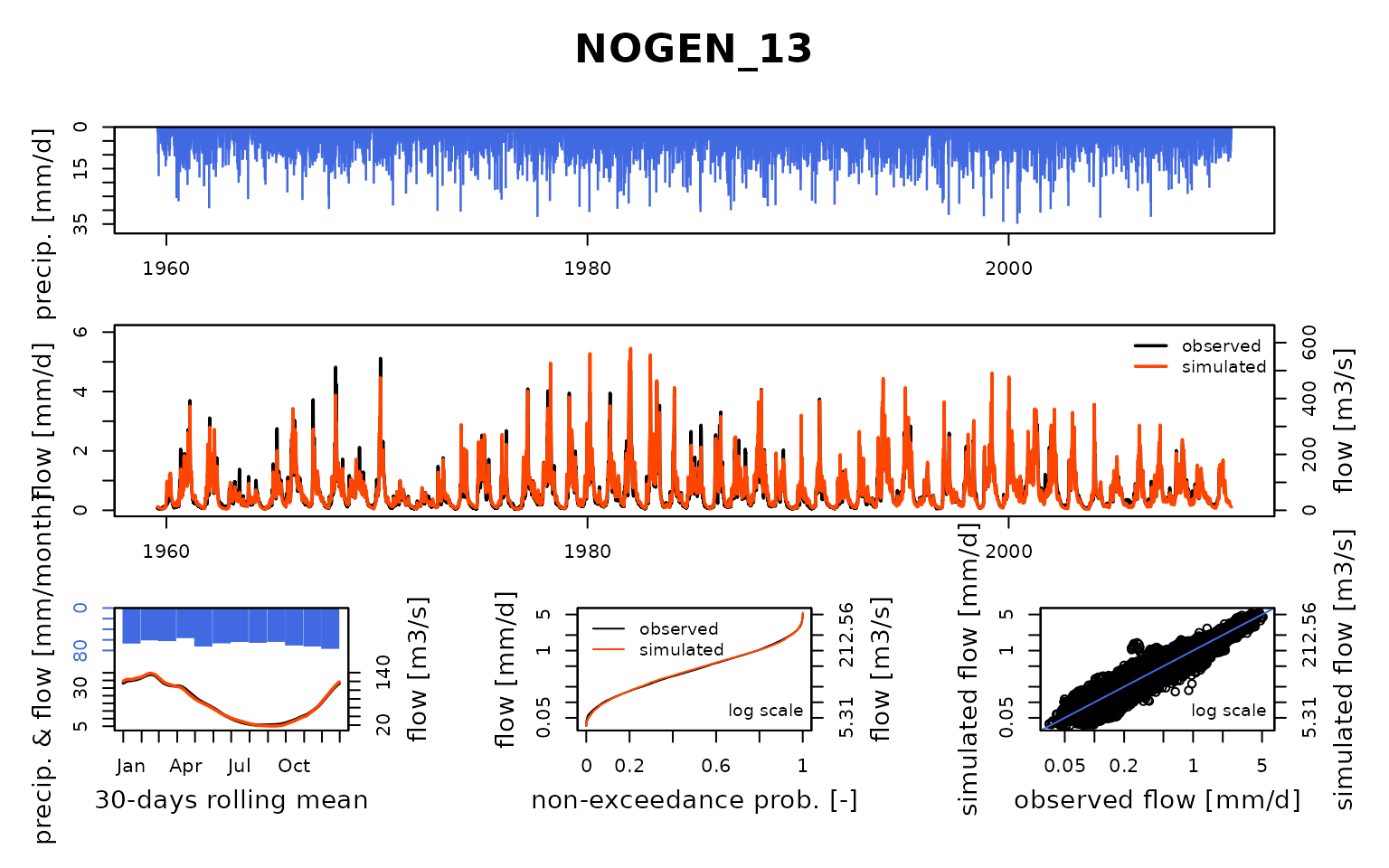
## Calibration.GRiwrmInputsModel: Processing sub-basin 'NOGEN_13'...
## Parameter regularization: test a priori parameters from node MERY-_22: 0.33, 749.945, -3.589, 60.34, 4.75
## Crit. KGE2[Q] = 0.9516
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9582
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.0104
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.0221
##
## Parameter regularization: test a priori parameters from node ARCIS_24: 0.32, 539.153, -2.973, 81.451, 5.244
## Crit. KGE2[Q] = 0.9398
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9591
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.0150
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.0416
##
## Parameter regularization: set a priori parameters from node MERY-_22: 0.33, 749.945, -3.589, 60.34, 4.75
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (243 runs)
## Param = 1.250, 432.681, -2.376, 83.096, 2.384
## Crit. Composite = 0.9298
## Steepest-descent local search in progress
## Calibration completed (31 iterations, 531 runs)
## Param = 0.240, 796.319, -3.780, 59.145, 4.745
## Crit. Composite = 0.9674
## Formula: sum(0.86 * KGE2[sqrt(Q)], 0.14 * GAPX[ParamT])
## Calibration.GRiwrmInputsModel: Processing sub-basin 'NOISI_17'...
## Parameter regularization: test a priori parameters from node MONTR_18: 1, 292.949, -0.578, 40.854, 3.355
## Crit. KGE2[Q] = 0.7547
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9104
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.0919
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.2090
##
## Parameter regularization: test a priori parameters from node CHALO_21: 0.42, 601.845, -2.06, 113.296, 6.546
## Crit. KGE2[Q] = 0.8469
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9334
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.0034
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.1378
##
## Parameter regularization: set a priori parameters from node CHALO_21: 0.42, 601.845, -2.06, 113.296, 6.546
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (243 runs)
## Param = 1.250, 432.681, -2.376, 83.096, 2.384
## Crit. Composite = 0.8679
## Steepest-descent local search in progress
## Calibration completed (38 iterations, 602 runs)
## Param = 0.620, 1718.622, -5.460, 69.649, 6.458
## Crit. Composite = 0.9574
## Formula: sum(0.87 * KGE2[sqrt(Q)], 0.13 * GAPX[ParamT])
## Calibration.GRiwrmInputsModel: Processing sub-basin 'COURL_23'...
## Parameter regularization: test a priori parameters from node GURGY_02: 1.422, 520.044, -2.61, 100.23, 3.976
## Crit. KGE2[Q] = 0.9584
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9656
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.0205
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 0.9888
##
## Parameter regularization: test a priori parameters from node BRIEN_03: 0.54, 219.203, -0.432, 81.451, 3.299
## Crit. KGE2[Q] = 0.8934
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9573
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.0607
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.0765
##
## Parameter regularization: test a priori parameters from node CHABL_12: 0.318, 195.523, -2.557, 34.22, 4.763
## Crit. KGE2[Q] = 0.8870
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9308
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 1.0868
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 0.9788
##
## Parameter regularization: set a priori parameters from node GURGY_02: 1.422, 520.044, -2.61, 100.23, 3.976
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (243 runs)
## Param = 1.250, 432.681, -0.020, 83.096, 2.384
## Crit. Composite = 0.9062
## Steepest-descent local search in progress
## Calibration completed (48 iterations, 705 runs)
## Param = 1.495, 960.102, -2.340, 175.144, 3.966
## Crit. Composite = 0.9629
## Formula: sum(0.86 * KGE2[sqrt(Q)], 0.14 * GAPX[ParamT])
## Calibration.GRiwrmInputsModel: Processing sub-basin 'MONTE_15'...
## Parameter regularization: test a priori parameters from node NOGEN_13: 0.24, 796.319, -3.78, 59.145, 4.418
## Crit. KGE2[Q] = 0.9530
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9745
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 0.9794
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.0336
##
## Parameter regularization: test a priori parameters from node COURL_23: 1.495, 960.102, -2.34, 175.144, 3.181
## Crit. KGE2[Q] = 0.9392
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9727
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 0.9775
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.0495
##
## Parameter regularization: set a priori parameters from node NOGEN_13: 0.24, 796.319, -3.78, 59.145, 4.418
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (243 runs)
## Param = 1.250, 432.681, -2.376, 83.096, 2.384
## Crit. Composite = 0.9475
## Steepest-descent local search in progress
## Calibration completed (26 iterations, 481 runs)
## Param = 0.250, 804.322, -3.780, 59.145, 4.414
## Crit. Composite = 0.9722
## Formula: sum(0.86 * KGE2[sqrt(Q)], 0.14 * GAPX[ParamT])
## Calibration.GRiwrmInputsModel: Processing sub-basin 'ALFOR_16'...
## Parameter regularization: test a priori parameters from node EPISY_14: 1, 610.285, -0.593, 41.294, 4.195
## Crit. KGE2[Q] = 0.8976
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9759
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 0.9922
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.0992
##
## Parameter regularization: test a priori parameters from node MONTE_15: 0.25, 804.322, -3.78, 59.145, 6.819
## Crit. KGE2[Q] = 0.9195
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9297
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 0.9886
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.0374
##
## Parameter regularization: set a priori parameters from node MONTE_15: 0.25, 804.322, -3.78, 59.145, 6.819
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (243 runs)
## Param = 1.250, 432.681, -2.376, 83.096, 2.384
## Crit. Composite = 0.9048
## Steepest-descent local search in progress
## Calibration completed (39 iterations, 612 runs)
## Param = 1.027, 2530.297, -4.825, 51.309, 6.754
## Crit. Composite = 0.9616
## Formula: sum(0.86 * KGE2[sqrt(Q)], 0.14 * GAPX[ParamT])
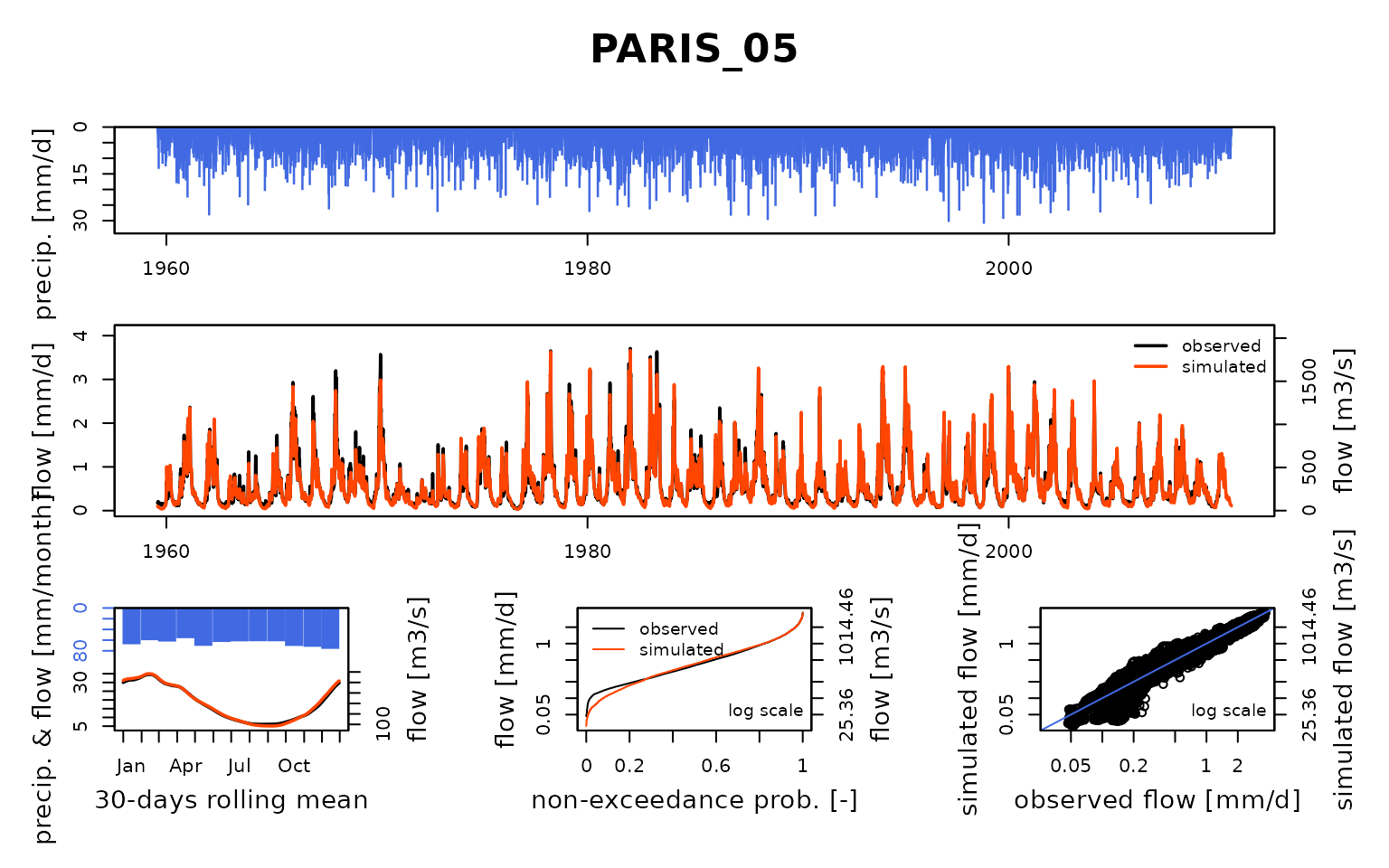
## Calibration.GRiwrmInputsModel: Processing sub-basin 'PARIS_05'...
## Parameter regularization: test a priori parameters from node NOISI_17: 0.62, 1718.622, -5.46, 69.649, 3.208
## Crit. KGE2[Q] = 0.9420
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9752
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 0.9656
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.0397
##
## Parameter regularization: test a priori parameters from node ALFOR_16: 1.027, 2530.297, -4.825, 51.309, 3.245
## Crit. KGE2[Q] = 0.9421
## SubCrit. KGE2[Q] cor(sim, obs, "pearson") = 0.9758
## SubCrit. KGE2[Q] cv(sim)/cv(obs) = 0.9653
## SubCrit. KGE2[Q] mean(sim)/mean(obs) = 1.0395
##
## Parameter regularization: set a priori parameters from node ALFOR_16: 1.027, 2530.297, -4.825, 51.309, 3.245
## Grid-Screening in progress (0% 20% 40% 60% 80% 100%)
## Screening completed (243 runs)
## Param = 1.250, 432.681, -2.376, 42.098, 2.384
## Crit. Composite = 0.9573
## Steepest-descent local search in progress
## Calibration completed (27 iterations, 492 runs)
## Param = 1.030, 2540.205, -4.837, 51.419, 3.242
## Crit. Composite = 0.9723
## Formula: sum(0.86 * KGE2[sqrt(Q)], 0.14 * GAPX[ParamT])Run the GR4J model with parameters obtained from the Michel calibration
Now that the model is calibrated, we can run it with the optimized parameter values:
ParamMichel <- extractParam(OutputsCalib)
OutputsModels <- RunModel(
InputsModel,
RunOptions = RunOptions,
Param = ParamMichel
)## RunModel.GRiwrmInputsModel: Processing sub-basin TRANN_01...## RunModel.GRiwrmInputsModel: Processing sub-basin STDIZ_04...## RunModel.GRiwrmInputsModel: Processing sub-basin BAR-S_06...## RunModel.GRiwrmInputsModel: Processing sub-basin CHAUM_07...## RunModel.GRiwrmInputsModel: Processing sub-basin CUSSY_08...## RunModel.GRiwrmInputsModel: Processing sub-basin STGER_09...## RunModel.GRiwrmInputsModel: Processing sub-basin GUILL_10...## RunModel.GRiwrmInputsModel: Processing sub-basin AISY-_11...## RunModel.GRiwrmInputsModel: Processing sub-basin EPISY_14...## RunModel.GRiwrmInputsModel: Processing sub-basin MONTR_18...## RunModel.GRiwrmInputsModel: Processing sub-basin LOUVE_19...## RunModel.GRiwrmInputsModel: Processing sub-basin LASSI_20...## RunModel.GRiwrmInputsModel: Processing sub-basin VITRY_25...## RunModel.GRiwrmInputsModel: Processing sub-basin GURGY_02...## RunModel.GRiwrmInputsModel: Processing sub-basin BRIEN_03...## RunModel.GRiwrmInputsModel: Processing sub-basin CHABL_12...## RunModel.GRiwrmInputsModel: Processing sub-basin CHALO_21...## RunModel.GRiwrmInputsModel: Processing sub-basin MERY-_22...## RunModel.GRiwrmInputsModel: Processing sub-basin ARCIS_24...## RunModel.GRiwrmInputsModel: Processing sub-basin NOGEN_13...## RunModel.GRiwrmInputsModel: Processing sub-basin NOISI_17...## RunModel.GRiwrmInputsModel: Processing sub-basin COURL_23...## RunModel.GRiwrmInputsModel: Processing sub-basin MONTE_15...## RunModel.GRiwrmInputsModel: Processing sub-basin ALFOR_16...## RunModel.GRiwrmInputsModel: Processing sub-basin PARIS_05...Save calibration data for next vignettes
save(ParamMichel, file = "_cache/V03.RData")