Web API
ODAM: Web API¶
Services¶
The ODAM API services are based on REST services using a Resource Naming convention i.e an understandable naming of resources leading to a Web API (resource identification/query) that is easily exploitable and easy to implement in a script language (R or Python).
Moreover, The ODAM API services follow the OpenAPI specifications.
API Documentation on SwaggerHub : INRA-PMB/ODAM/1.2.0
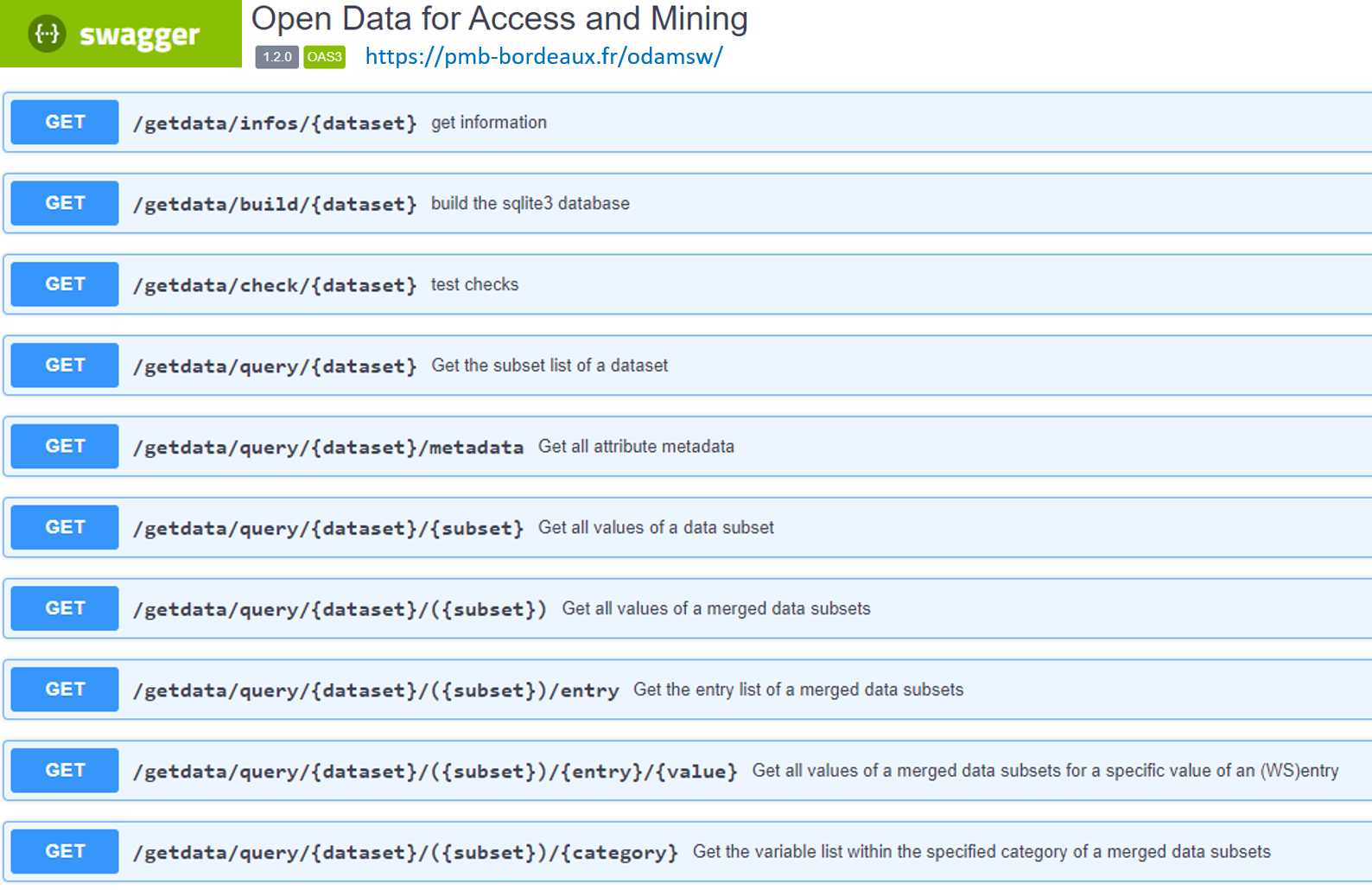
Four main services (i.e query-paths) are provided, as described below:
| Service | Description |
|---|---|
| check | Check the structure and consistency of the metadata files with the data files |
| build | Build the sqlite3 database allowing an acceleration of the queries |
| query | Query data based on metadata |
| infos | Get the content of the infos.md file (markdown format) |
-
query
Using the two metadata files namely a_attributes.tsv and s_subsets.tsv, it is possible to build a tree structure from which the data files can be queried to extract a subset. The tree structure is built on the Entity-Attribute-Value scheme.
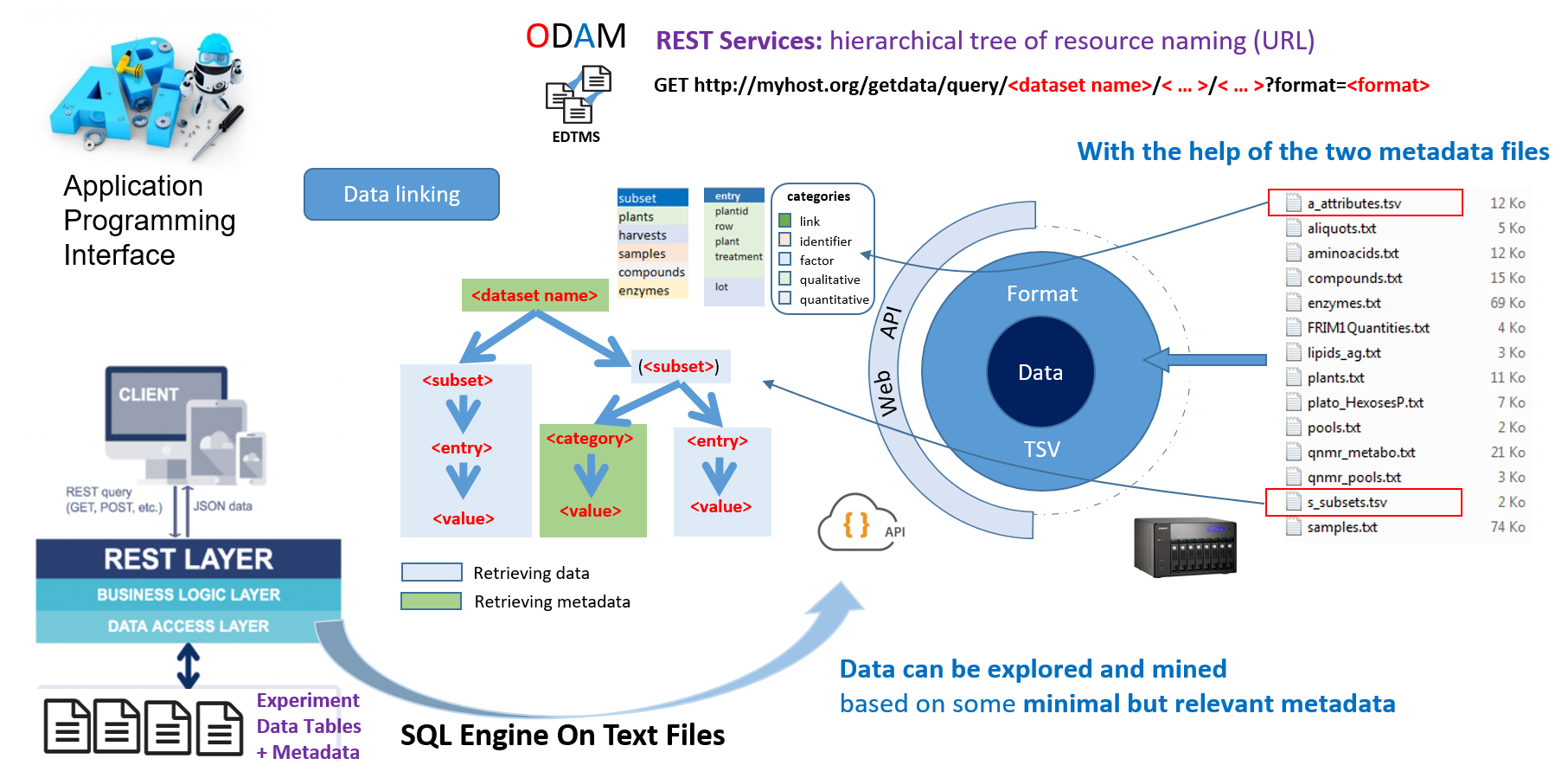
| Field | Description | Examples |
|---|---|---|
| <dataset name> | Short name (tag) of your dataset | frim1 |
| <subset> | Short name of a data subset | samples |
| <entry> | Name of an attribute entry (defined by the user in the a_attribute file (column ‘entry’) | sampleid |
| <category> | Name of the attribute category; (assigned by the user in the a_attribute file (column ‘category’). Possible values are: ‘identifier’, ‘factor’, ‘qualitative’, ‘quantitative’ | quantitative |
| (<subset>) | Set of data subsets by merging all the subsets with lower rank than the specified subset and following the pathway defined by the "obtainedFrom" links. | (samples) <=> plants + samples |
| <value> | Exact value of the desired entry or category | 1 (for subset) or Factor (for category) |
| <data format> | Format of the retrieved data; possible values are: CSV, TSV, JSON or XML | tsv |
-
infos
This service allows you to retrieve additional information. See Additional information in Data collection an preparation.
-
check
This service allows to perform a set of tests to check if the dataset is well formatted and structured according to the expected specifications. See Final checking in Data collection an preparation.
-
build
This service builds the corresponding sqlite3 database to speed up response times on large files. See below in the section Very detailed example of API querying.
Two additional services (i.e query-paths) are provided (see Additional information section in Data collection and preparation), as described below:
| Service | Description |
|---|---|
| image | Retrieve an image: /getdata/image/{dataset}/{imagefile.png} |
| Retrieve a PDF file: /getdata/pdf/{dataset}/{pdffile.pdf} |
Some services (i.e query-paths) are available as shortcuts
| Service | Description |
|---|---|
| json | Query data with data formatted in JSON as output (shortcut for ../query/{dataset}/...?format=json) |
| tsv | Query data with data formatted in TSV as output (shortcut for ../query/{dataset}/...?format=tsv) |
| xml | Query data with data formatted in XML as output (shortcut for ../query/{dataset}/...?format=xml) |
Parameters that can be specified in the query-string as described below. These parameters can be combined : e.g ?format=tsv&limit=100&debug=1
| Parameter | Description | Example |
|---|---|---|
| format | Format of the retrieved data; possible values are: 'xml', ‘json’, 'csv' or ‘tsv' | format=tsv |
| auth | Set the API Key if required (non-preferential method) | auth=secret.key |
| limit | Limitation of the number of records provided | limit=100 |
| debug | Provide information about the request (1 for debug, 0 for query output) | debug=1 |
Authorization mechanism¶
Currently, the ODAM API implements a key-based authorization mechanism, which is different from the authentication mechanism. (see this blog).
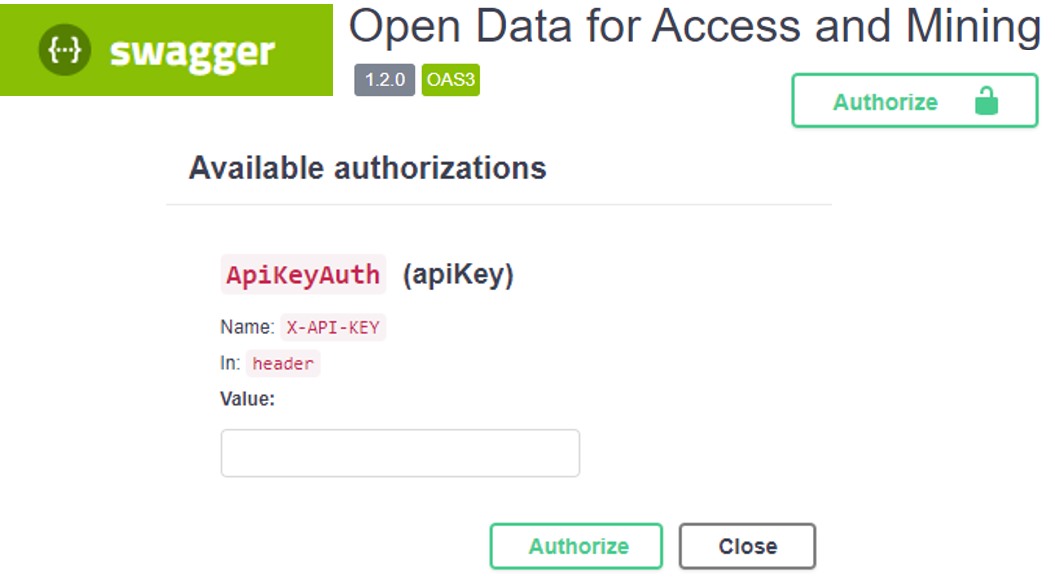
The API key approach seems to us to be an adequate solution insofar as only the "read" functionality is concerned. Thus, without the need to edit, modify or delete, security is less of a concern. Furthermore, we have associated to these API keys the possibility to limit their usage according to the internet address (IP) of the machines from which the users make their requests.
There is no centralized management of these keys. The management is done for each dataset independently. The principle is simple: if you want to put a key protection on a dataset, just create a file named authkeys.tsv (TSV format) and add it in the same directory as the dataset. Without this file, the dataset is completely open.
- The file authkeys.tsv (TSV format) has three columns as described below:
| column | description |
|---|---|
| 1st | IP Address or sub-network : in case of sub-network, only the common part of all IP adresses in the sub-network have to be specified. To signify all IPs, the character '*' will be used |
| 2nd | Can be either the API key or one of the two following keywords: 'deny' or 'allow'. 'deny' to forbid access (with or without API key), and 'allow' to allow access without API key. |
| 3rd | Comment or description |
Knowing that as soon as a rule is satisfied, the other rules are not read, it is important to order the rules according to a decreasing priority, the first one being the most important and so on.
- Example of Authorization file (authkeys.tsv)
| IP | Authorization | Description |
|---|---|---|
| 77.111 | deny | VPN Opera Browser |
| 10.0.0 | allow | local VMs |
| 192.168.100.50 | allow | a PC Desktop |
| 192.168.100.51 | allow | another one PC Desktop |
| * | secret.key | Others |
- Another example of authorization file for a private dataset could be this:
| IP | Authorization | Description |
|---|---|---|
| 192.168.100.50 | allow | a PC Desktop |
| * | deny | Others |
-
The API key, when required, can be specified:
- in the API URL by adding 'auth={API key}' as a query parameter (i.e query string) :
curl "https:/myserver/{query-paths}/{dataset}/.../?format={format}&auth={API key}"- in the HTTP header (recommended method):
curl "https:/myserver/{query-paths}/{dataset}/.../?format={format}" -H "X-Api-Key: {API key}"
Note 1: You have to take into account that there may be several network cards on your machine, especially virtual network cards in the case of using a virtualization software (e.g. VirtualBox). So it is the IP address of the virtual card involved that should be put in the authkeys.tsv file in order to set specific authorizations. For example, the virtual network card of the distributed virtual machine for VirtualBox (see Installation) has the IP address set as 192.168.99.100. See ipconfig for Windows and ifconfig/ip for linux.
Note 2: If an application hosted on a remote server makes requests, then of course the IP address of that server (which is not the IP of the client) will be taken as the default for comparison with the authorization settings. However, it is possible to implement a mechanism to modify the API query header by specifying the client's originating IP (see X-Forwarded-For). This is what has been done for example in the data explorer.
The approach used in the data explorer is as follows:
- using a javascript, we retrieve the client's IP address which is sent to the server side (see examples)
- On the server side, we modify the API request header by adding the X-Forwarded-For parameter positioned with the client IP address (using the R package httr, see add_headers)
Very detailed example of API querying¶
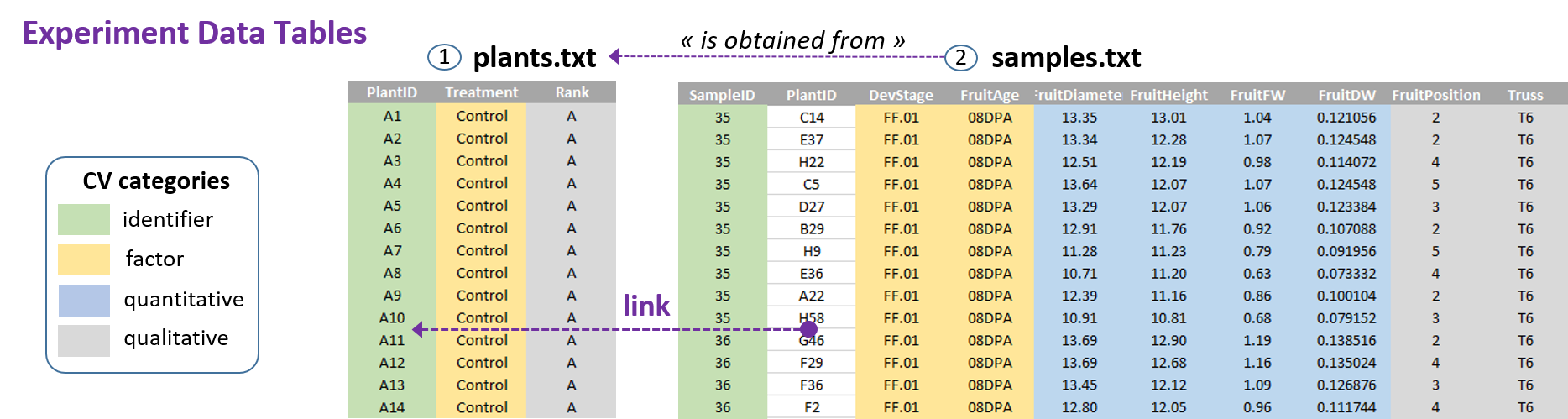
Since all the experimental data tables were generated as part of an experiment associated with a Design of Experiment (DoE), each file thus contains data acquired sequentially as the experiment progressed. There must therefore be a link between each file, i.e. information that connects them together. In most cases (if not all), this information corresponds to identifiers that make it possible to precisely reference within the experiment each of the elements belonging to the same observation entity forming a coherent observation unit. For example, each plant, each sample has its own identifier, and each of these entities corresponds to a separate data file.
The files generated during data collection have to be organized according to the entity-relationship model similar to relational database management systems (RDBMS), as shown below with the FRIM1 dataset.
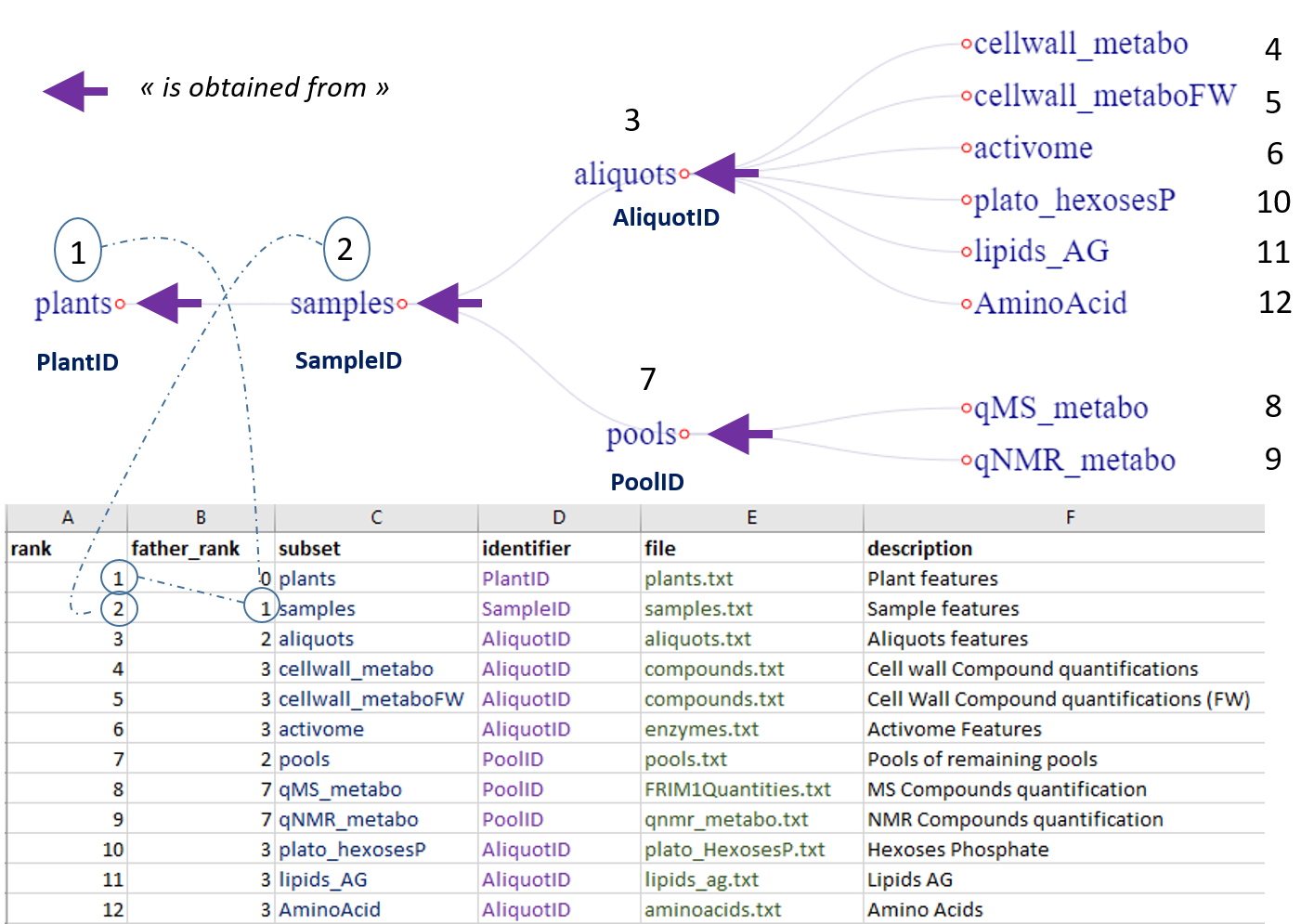
Extract of structural metadata (file s_subsets.tsv) for FRIM1 dataset
Each file (an entity, i.e an observational unit) corresponds to a type of collected data (samples, compounds, ...) for which is associated a set of attributes, i.e. a set of variables that may include observed or measured variables (quantitative or qualitative), controlled independent variables (factors) and obviously an identifier.
Since there is a relationship between each of the files, it is therefore possible to combine them by following the paths of their mutual links. Suppose we want to retrieve activome data (enzyme expressions) combined with NMR observed metabolite data, including experimental metadata (factors and phenotypic data).
The ODAM API offers the possibility of combining data by following relational paths, using the '({subset})' operator in the API query. Simply specify the desired subsets separated by a comma, as shown below:
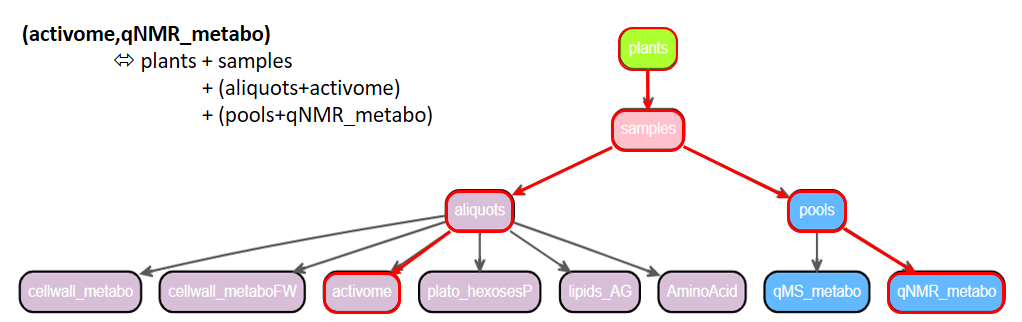
'(activome,qNMR_metabo)' operator in the API query
The '({subset})' operator allows you to get data subsets by merging all the subsets with lower rank than the specified subsets and following the pathway defined by the “obtainedFrom" links.
Let's further assume that we only want the data for the 'Control' treatment. The ODAM API also offers the possibility of setting a selector called an 'entry' provided that such an entry has been specified for the attribute to which one wants to apply a selection, as shown below:
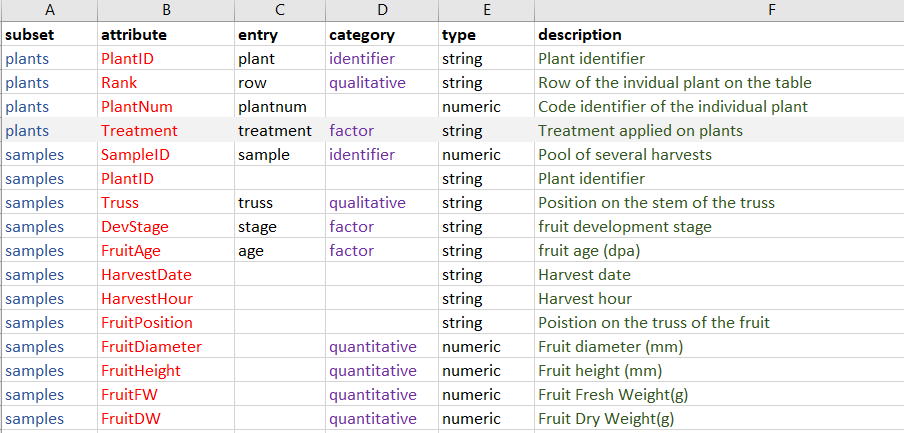
Extract of structural metadata (file a_attributes.tsv) for FRIM1 dataset
In our case, the 'Treatment' factor has the entry (alias) 'treatment'. It is therefore possible to use it as a selector in the API query. Thus, our complete API query would be this one:

Complete API query
To test this query by yourself, you can go to ODAM's Swagger interface and play with the followed specific query :

ODAM API in Swagger
Finally, how does it work internally? How are the different subsets of data combined in practice? Since each data subset being a data file, and linked to one or more other data subsets by means of common identifiers, it is possible to apply a SQL query from the data files as shown below:

SQL query corresponding to the API query
The application of SQL queries is performed
-
either with the q - Text as Data tool which allows to apply SQL queries directly on CSV and TSV files. It is the default tool.
-
or with the sqlite3 tool which, by building a database and indexing it, allows to accelerate considerably the response times on large files. To proceed with the creation and indexing of the database, a call to the API is necessary (/getdata/build/{dataset}). To see what types of operations are made, try i.e this API query
In addition, if you want to display information about the API query, just add '/debug' at the end of the query, as shown below:
https://pmb-bordeaux.fr/getdata/query/frim1/(activome,qNMR_metabo)/treatment/Control/debug