Presentation
ODAM: Presentation¶
ODAM (Open Data for Access and Mining) is an Experiment Data Table Management System (EDTMS)
Background¶
In life sciences, (and particularly in plant sciences), each time an experimental design (DoE) is implemented, we can, very schematically, represent the data flow according to 4 main steps, from raw data to publication of results.
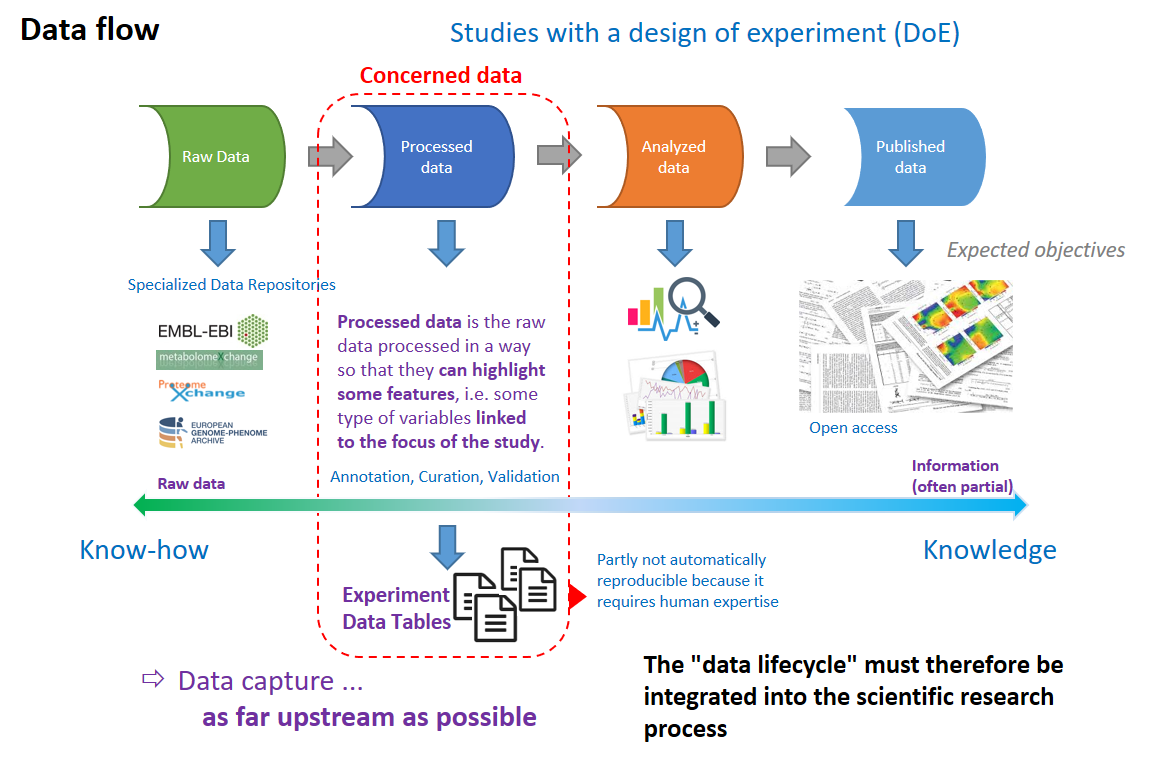
Very Schematically one can represent the data flow according to 4 main steps, from raw data to publication of results.
- What kind of data
From a biological point of view, the data integrating the maximum amount of relevant information are those resulting from the pre-processing of so-called raw data (resulting from analytical techniques) and including annotations with curation then validated by one or more experts; i.e. those involving a transformation of analytical variables (peaks or resonances on spectra, locus on a DNA / RNA / Protein sequence, ...) into biological variables (metabolites, proteins, genes, ...). At this stage, because they are not synthesized, they still have all their variabilities (technological and biological replicas on all factorial levels) and therefore have more potential for reuse. Moreover, these data are not automatically reproducible (e.g. via workflows) because they require human expertise (i.e. know-how). This is why we have focused our data management on these experimental data tables.
- Data handling
When generating data in an experiment involving several types of data from several analytical techniques, and this for the same samples, the task of being able to easily link these different data on the basis of sample identifiers is crucial. Indeed, as data consistency must be ensured throughout the study, it should be ensured that it is not necessary to conduct a laborious survey to find the right identifiers, but to rely confidently on a data management system.
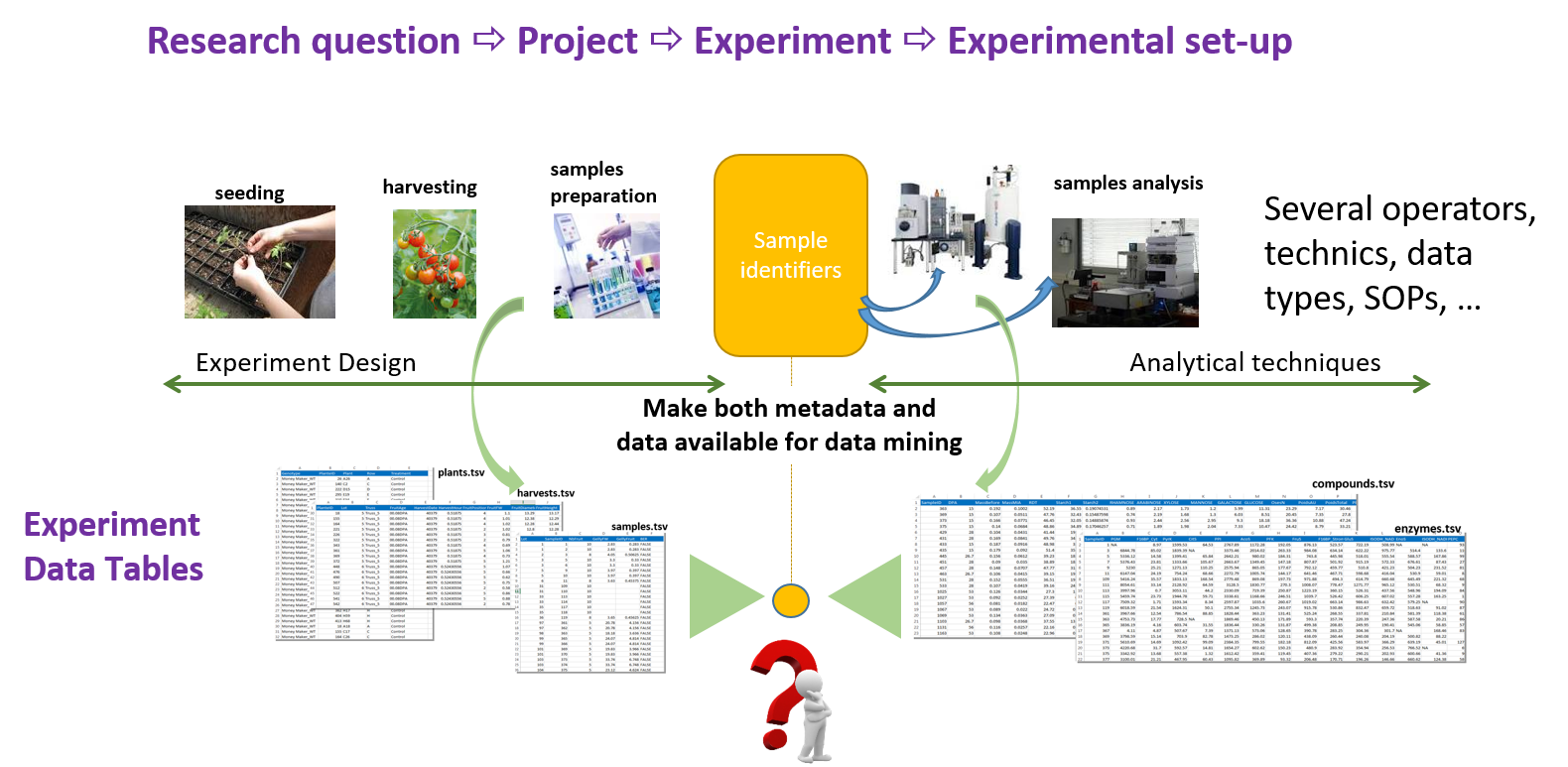
Linking each analysis to its corresponding sample throughout the study without laborious investigation or data handling is crucial to ensure data consistency.
- Data sharing
Each time we plan to share data coming from a common experimental design, the classical challenges for fast using data by every partner are data storage and data access. We propose an approach for sharing project data all along its development phase, from the setup of the experimental schema up to the data acquisition from the various analyzes of samples, so that all data is readily available as soon as they are generated.
Proposed solution¶
ODAM software is designed to manage experimental data tables in a quick and easy way for users. There is no need to develop a complex data model. Just complete the data with some structural metadata. These structural metadata will be used first to make full use of the data as soon as they are produced and formatted and then to annotate the dataset for later dissemination, either to project partners or more widely.
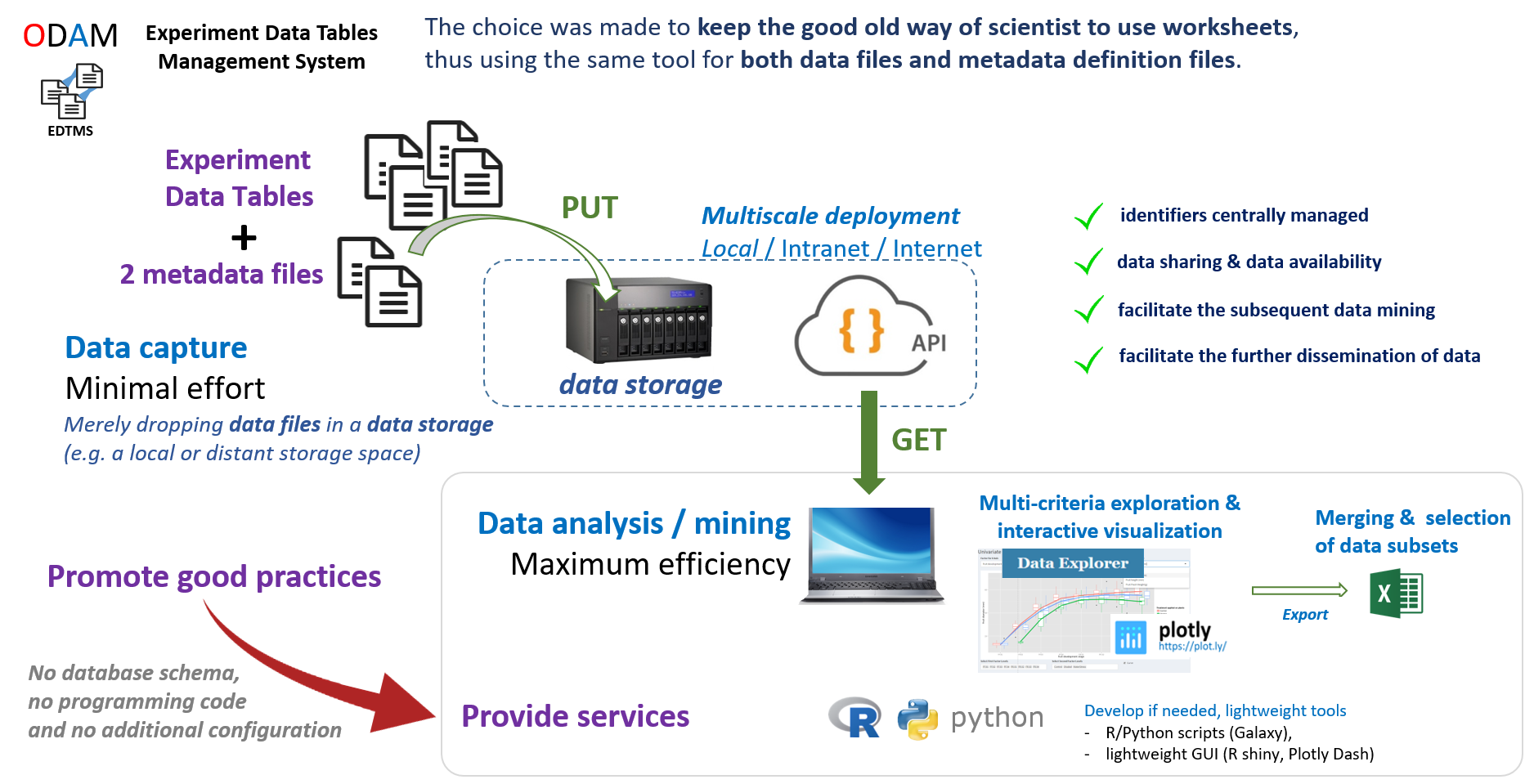
The core idea in one shot
The central idea which has been the founding idea of ODAM, is that data producers "just" have to drag and drop their data tables onto a storage space, which depending on the chosen infrastructure can be local (i.e. their PC, or a NAS) or remote (virtual disk space). So simply dropping data files (along with two additional metadata files) on the storage space allows users to access them through web services. This means there is no need for additional configuration on the server.
ODAM proposes to meet certain needs typically encountered during the implementation of an experimental design in life science including several different analyses of the same sample.
- Data collecting and preparation
The formatting of all the data and matching the data from the different analyses with their experimental context can be a long step. Tasks such as collecting and preparing data in order to combine several data sources require a lot of long, repetitive and tedious data handling. Similarly, when modeling, subsets must be selected and then many scenarios with different parameters must be tested. (e.g data scientists spend 79% of their time on data preparation1)
- Data sharing
Enabling centralized management of identifiers (e.g. plants, crops, samples, etc.) so that they are unique and shared by all project members. Indeed, as each biological sample is most often aliquoted and then sent for analysis by different techniques, the data returned in tabular form must be able to be linked to the other data according to the identifiers of the samples. Giving access to data for rapid use by each project member and this throughout the development phase, from the implementation of the experimental design to the acquisition of data from the various sample analyses, so that all data are readily available as soon as they are generated.
- Data publishing
-
- To be able to publish one's data without a colossal effort in data curation, and without the need for data archaeology.
- To be able to publish one's data according to the FAIR principles, at least the essentials.
- To facilitate the reuse of data by providing structural metadata, thus avoiding that data consumers spend a disproportionate amount of time trying to understand the digital resources they need and devising specific ways to combine them.
ODAM sofware suite allows experimental data tables to be widely accessible and fully reusable and this with minimal effort on the part of the data provider.
- Easily structure your data by adding structural metadata so that you can first exploit it locally yourself, before sharing it more widely just as easily.
- make research data locally or broadly accessible all along the project
- allow data to be selected then, downloadable by web API
- allow data and analysis to be visualized online
Based on the following criteria:
- Centralized management of identifiers (individuals, samples, aliquots, etc ...) so that they are unique and shared by all
- Avoid the implementation of a complex data management system (requiring a data model) given that many changes can occur during the project. (possibility of new analysis, new measures or give up some others, ...)
- Facilitates the subsequent publication of data: either the data can serve to fill in an existing database or the data can be broadcast through a web-service approach with the associated metadata.

ODAM can be seen as a wheel ...
Advantages of this approach
According to us the most important thing is to capture structural metadata as early as possible. The objective of our approach is to make this upstream capture an advantage to facilitate data analysis and therefore an incentive to perform this metadata capture. However on the basis of metadata files this does not prevent the development of parsers/converters to other formats or a connection to a more complex data model (e.g. ISA-TAB, MIAPPE 1.1) in order to include the data (transformed or not) in existing standards-compliant data infrastructures (e.g. SEEK platform)
Guideline for good data management
ODAM's general philosophy is to consider that the data life cycle must be integrated into the scientific research process. Furthermore, by considering the FAIR principles as a guide to produce data that is reproducible and frictionless reusable by humans and machines, data FAIRification process is accomplished without insurmountable effort (FAIR by design)
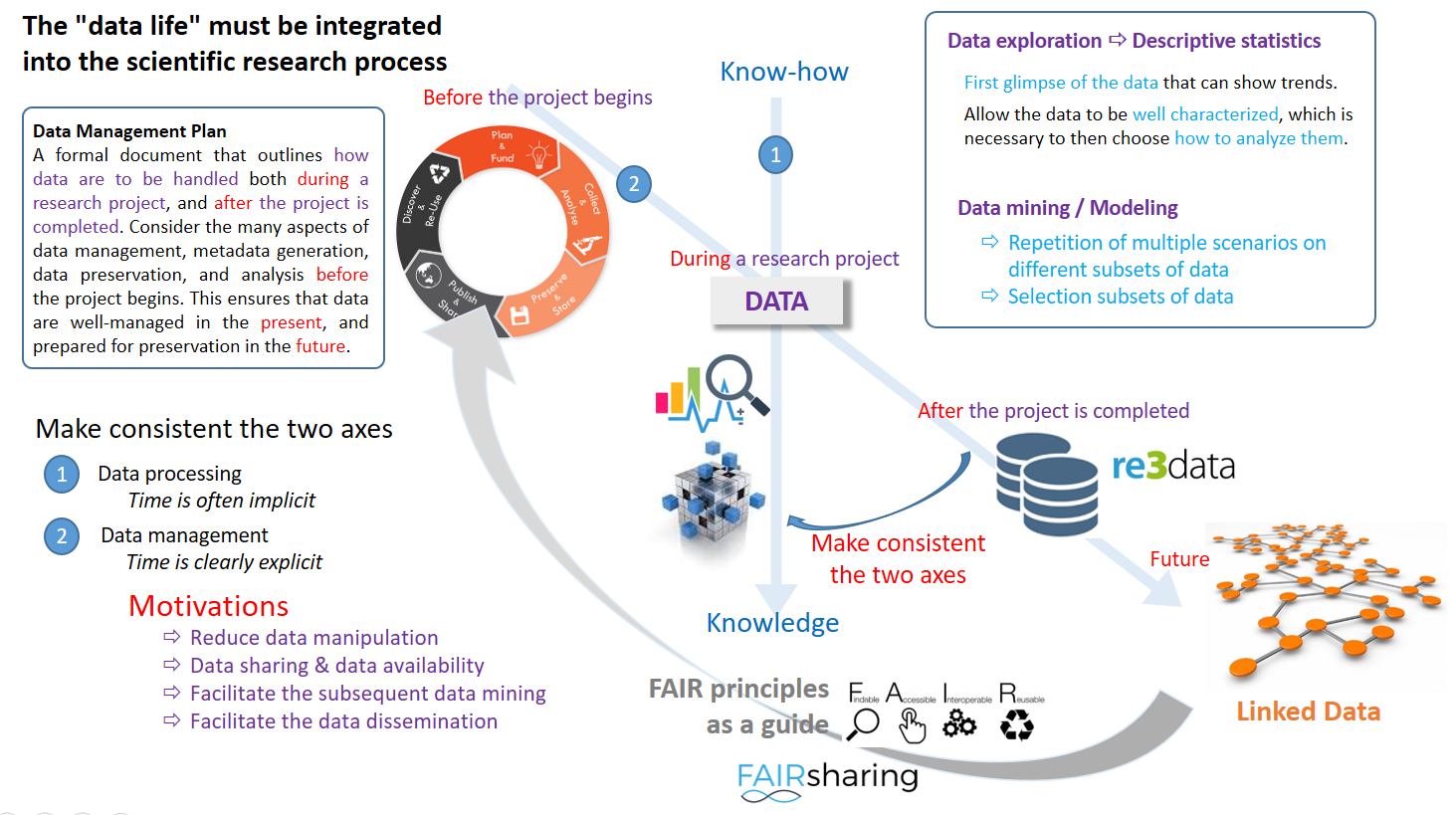
For this work, we made the choice to keep the good old way of scientist to use worksheets, thus using the same tool for both data files and metadata definition files.
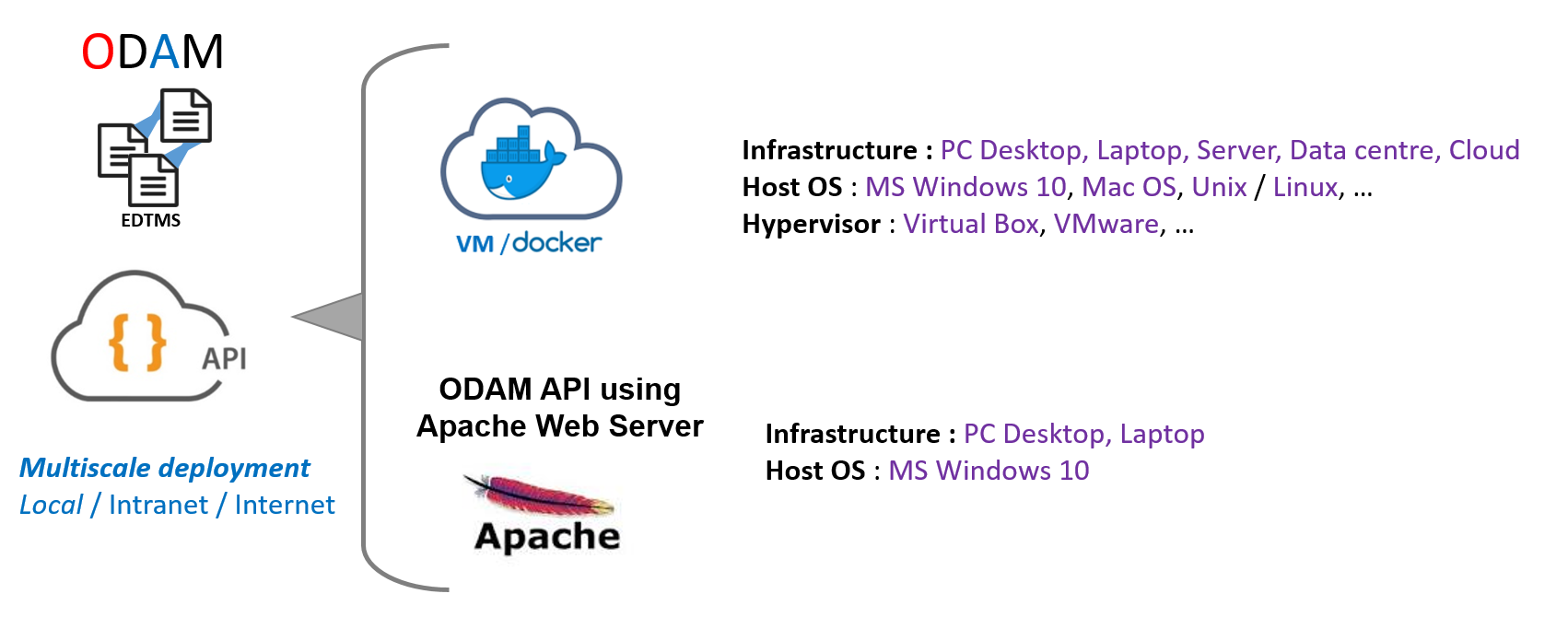
The ODAM software can be deployed at multiple scales (local, intranet, internet), depending on the need and the target community.
Guideline keywords : experimental data tables, Research Data Management, OpenAPI, FAIR Data principles, FAIR assessment
Documentation¶
- Presentation : A presentation on the ODAM software, its aims and what can we do with it for what purposes.
- Presentation : How to best manage your data to make the most of it for your research
- Presentation : A presentation about Data Management in the context of Open Science.
- Presentation : Rendre plus FAIR les tableaux de données expérimentales dans les sciences de la vie
-
CrowdFlower, Data Science Report (2016) https://visit.figure-eight.com/rs/416-ZBE-142/images/CrowdFlower_DataScienceReport_2016.pdf ↩