A dataset example
ODAM: A dataset example¶
The FRIM dataset¶
Fruit Integrative Modelling, an ERASysBio+ project : Yves Gibon (Coordinator)
The project aimed to build a virtual tomato fruit that enables the prediction of metabolite levels given genetic and environmental inputs, by an iterative process between laboratories which combine expertise in fruit biology, ecophysiology, theoretical and experimental biochemistry, and biotechnology.
Purposes of the project
To build a kinetic model encompassing the routes carbon takes, once imported into the fruit cells from the source organs of the mother plant.
To integrate the kinetic model with a phenomenological model predicting sugar and organic acid contents as functions of time, light intensity, temperature and water availability.
To obtain large-scale experimental measures of the consequences of altered environmental conditions.
To assess the influence of the environment on fruit metabolism, tomato (Solanum lycopersicum 'Moneymaker') plants were grown under contrasting conditions (optimal for commercial, shaded production) and locations. Samples were harvested at nine stages of development, and 36 enzyme activities of central metabolism were measured as well as protein, starch, and major metabolites, such as hexoses, sucrose, organic acids, and amino acids.
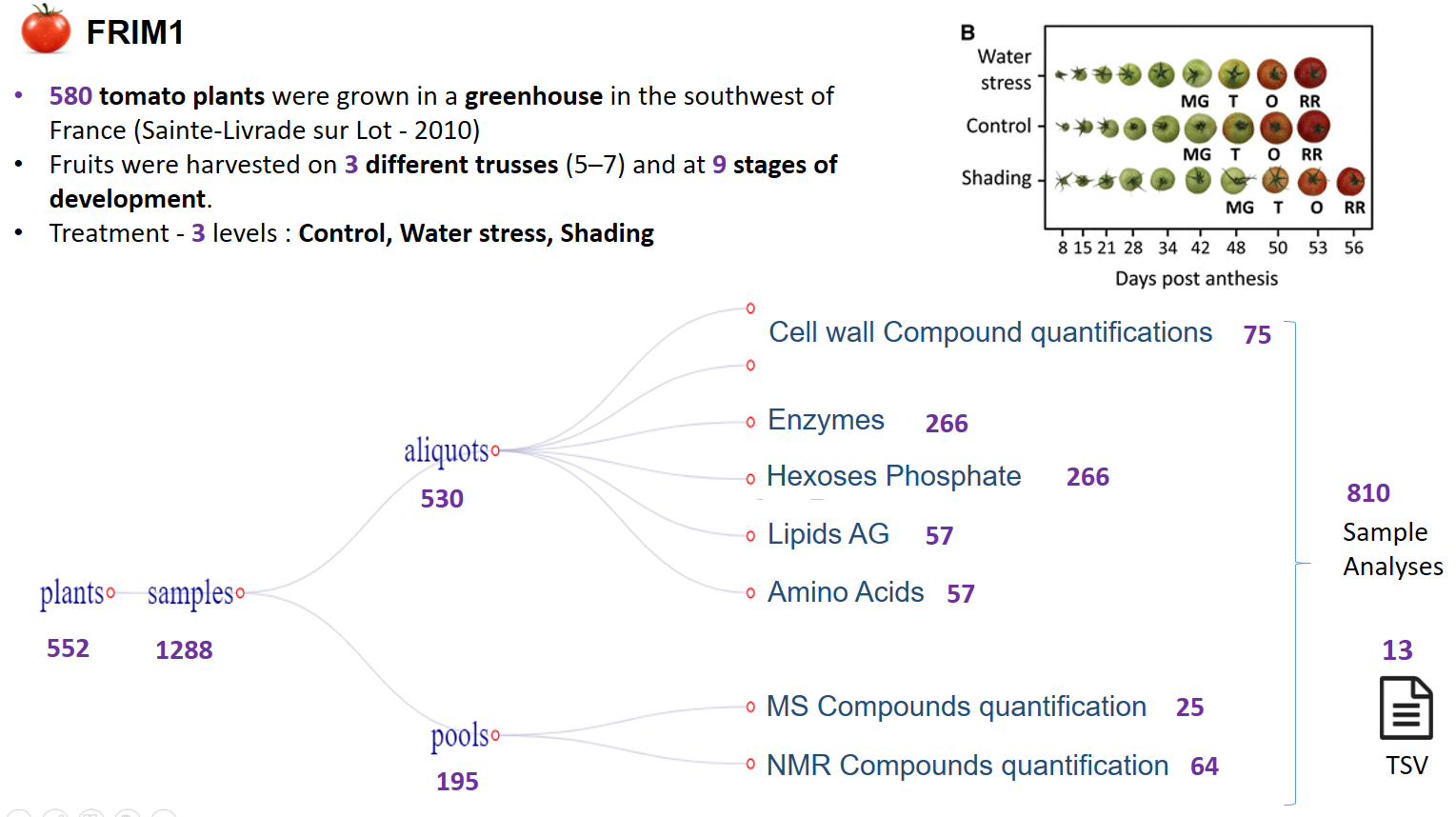
Experimental data tables from the Frim project ('frim1' study)
About 580 tomato plants were grown in a greenhouse in the southwest of France (Sainte-Livrade sur Lot) during the summer of 2010 according to usual production practices.
Links related to the dataset¶
| Description | Link |
|---|---|
| Data INRAE(*) | https://doi.org/10.15454/95JUTK |
| Data explorer(*) | https://pmb-bordeaux.fr/dataexplorer/?ds=frim1 |
| Modeling the growth of tomato fruits based on enzyme activity profiles : An example of data analysis interfaced by ODAM | https://hal-cnrs.archives-ouvertes.fr/hal-02611223/file/FRIM1_Growth_model.html |
| Jupyter notebooks (R & Python) | https://nbviewer.jupyter.org/github/djacob65/binder_odam/tree/master/ |
(*) Both repositories are supported by INRAE (France) for a minimum period of 10 years (until 2030)
Publication of the dataset according to FAIR principles¶
Data publishing
Because ODAM is primarily an Experimental Data Table Management System (EDTMS) for data sharing, it must be associated with a suitable data repository in order to support data publishing. So the ODAM approach has to be regarded as complementary with publication of the data online within an institutional data repository as described in re3data.org (e.g. Data INRAE) associated or not with a scientific paper.
To be compliant with the FAIR principles, not all data, documents, workflows and other tools need to be located in a single system, but from a central repository, it is the set of links that constitutes the true information management system. It must be able to be traversed by a human being as well as by machines.
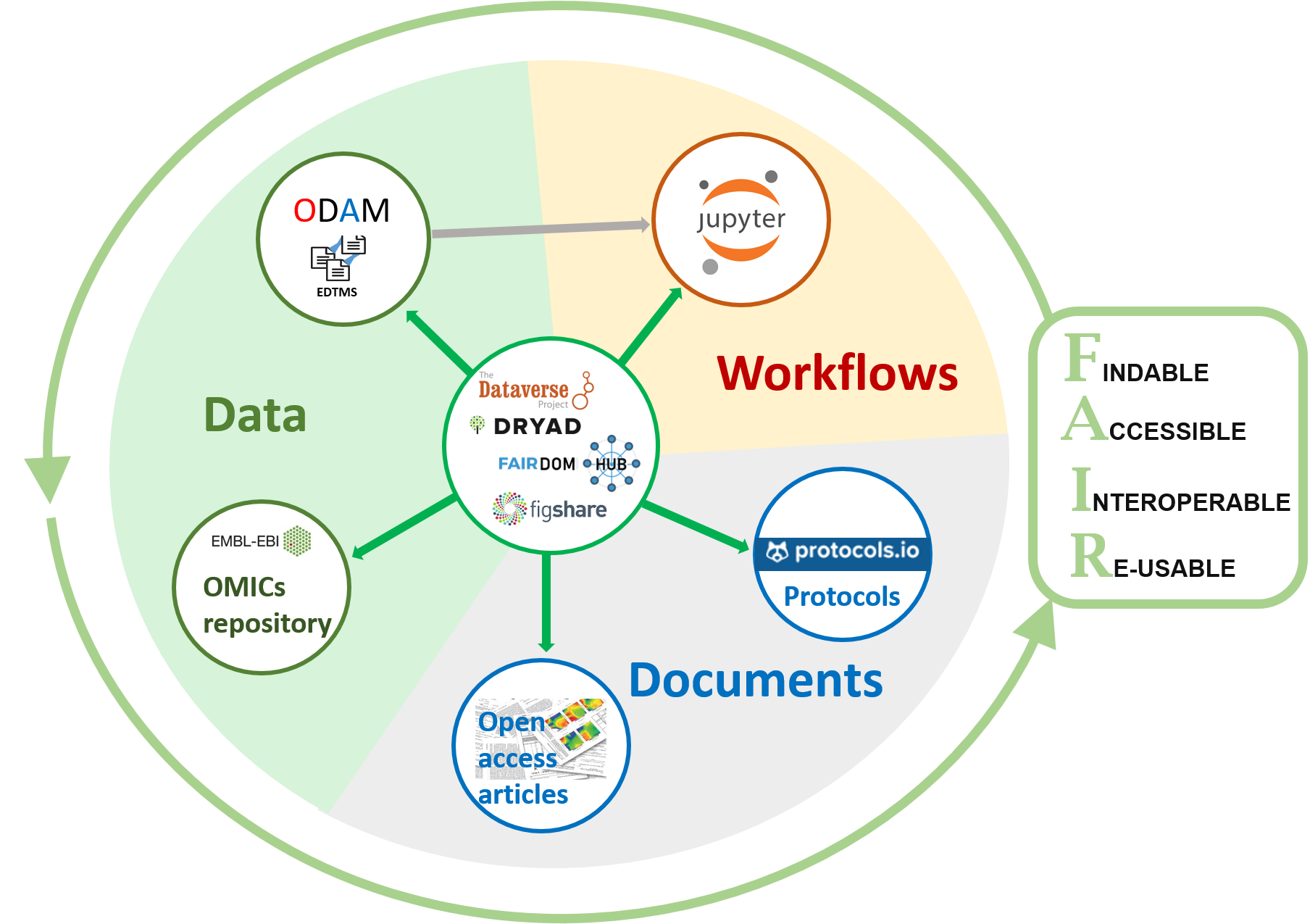
Dataset information need to be linked but not necessary in the same repository
Data standardisation
Data INRAE repository as a hub (based on Dataverse) allows to interconnect the different elements of the FRIM dataset. A file named 'datapackage.json', structured according to an explicit schema, was generated based on the structural metadata previously defined with the help of spreadsheets. By relying on explicit schemas (JSON-LD, JSON Schema) for both metadata and data, this makes it possible for both humans and machines to reuse data without friction..
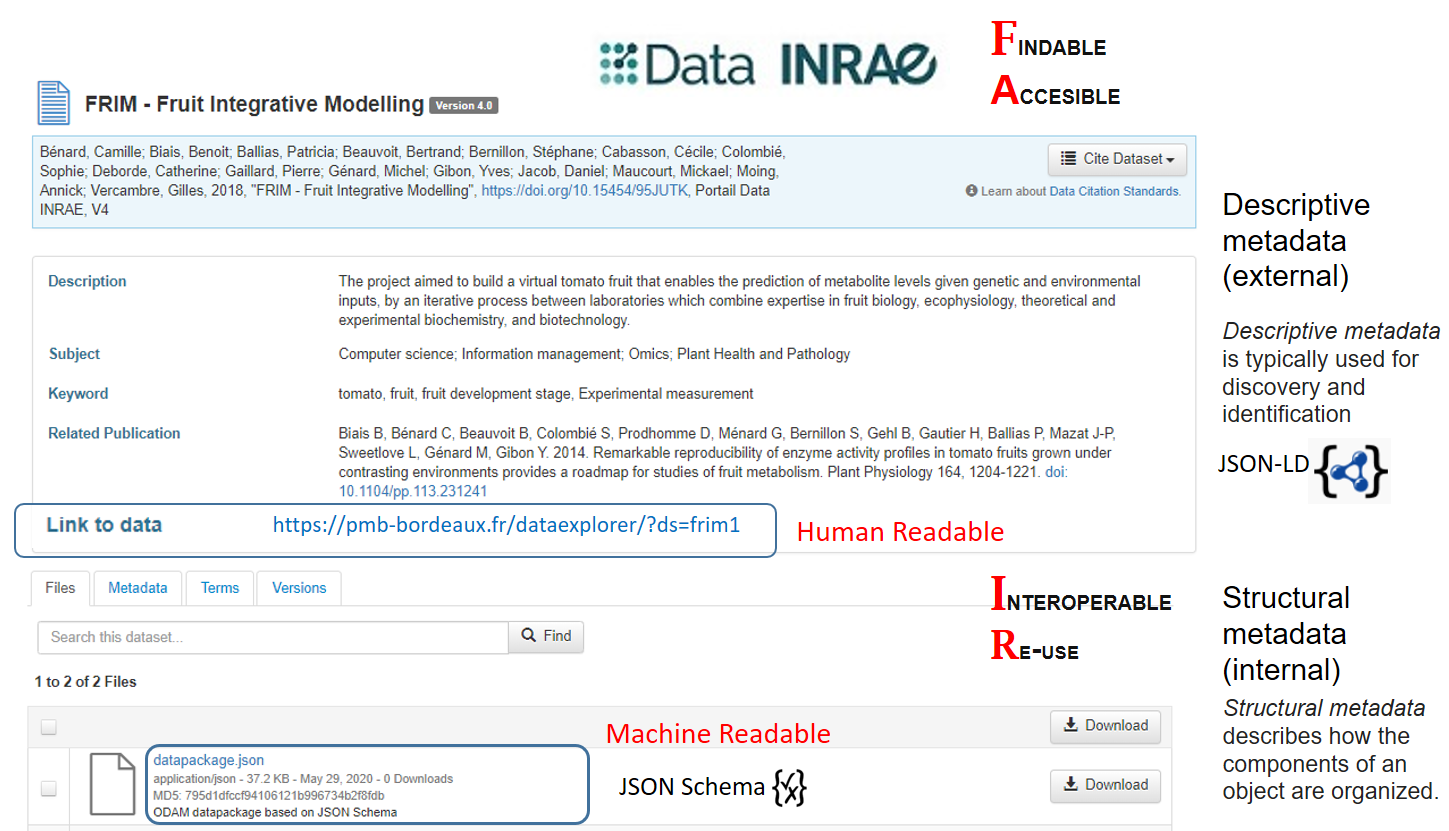
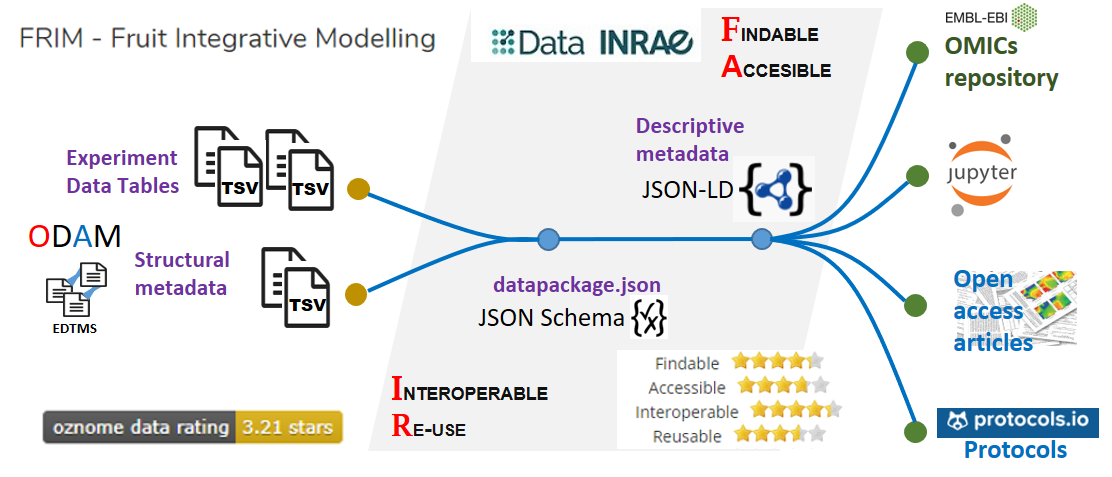
Data INRAE repository as a hub (based on Dataverse)
An explicit schema allows to define structural metadata along with unambiguous definitions of all internal elements (e.g. column definitions, units of measurement), through links to accessible (standard) definitions. Thus, this results in better annotated and more easily usable data that meets effortlessly the FAIR criteria for reusability. Indeed, structured data using a discoverable, community-endorsed schema or data model can only have a positive impact on the FAIR criteria 'Interoperable' and 'Reusable'.
Data referencing
In addition, a good practice for referencing datasets on the Internet is to manage URIs instead of URLs. A URI is a string of characters that unambiguously identifies a particular resource. It consists of a prefix linked to the resource and an identifier within the resource.
For this purpose, the ODAM repository was registered at the central registry Identifiers.org1. Its URI consists of the prefix 'odam' followed by the short name of the dataset, i.e. odam:<dataset>. The resource pointed to by this URI after de-reification is precisely the structural metadata returned according to the JSON datapackage schema.
URI of the Frim1 dataset: odam:frim1 which can be de-reified using the identifiers.org registry :
See also for more details:
- ODAM data-package based on JSON-Schema
- ODAM data-package from a Dataverse repository
- Find a Dataverse dataset via Google Search
- FAIR grids applied on FRIM dataset
References¶
- Biais B, Bénard C, Beauvoit B, Colombié S, Prodhomme D, Ménard G, Bernillon S, Gehl B, Gautier H, Ballias P, Mazat J-P, Sweetlove L, Génard M, Gibon Y. 2014. Remarkable reproducibility of enzyme activity profiles in tomato fruits grown under contrasting environments provides a roadmap for studies of fruit metabolism. Plant Physiology 164, 1204-1221. doi: 10.1104/pp.113.231241
-
Nick Juty, Nicolas Le Novère, Camille Laibe, Identifiers.org and MIRIAM Registry: community resources to provide persistent identification, Nucleic Acids Research, Volume 40, Issue D1, 1 January 2012, Pages D580–D586, https://doi.org/10.1093/nar/gkr1097 ↩
